Research Article - Biomedical Research (2017) Volume 28, Issue 20
Examining the relationship between MCPH1 with H2AX in esophageal cancer cells ECA109
Hongyun Shi1, Ke Zhang1, Ming Wen1 and Yu Yuan2*
1Department of Radiation Oncology, the Affiliated Hospital of Hebei University, Baoding, China
2Department of Neurosurgery, the Affiliated Hospital of Hebei University, Baoding, China
- *Corresponding Author:
- Yu Yuan
Department of Neurosurgery
The Affiliated Hospital of Hebei University, China
Accepted on October 24, 2017
Abstract
Objective: To discover the role of MCPH1 in the DNA double-strand damage induced by ionizing radiation and the relationship with H2AX in esophageal cancer cell ECA109.
Methods: ECA109 cancer cells will be accepted 8 Gy 1 h after irradiation for protein extraction and immunofluorescence observed MCPH1 and H2AX protein expression and nuclear foci changes. Establish a stable low expression of H2AX cell lines. Detecting MCPH1 and H2AX protein expression and nuclear foci changes induced by ionizing radiation after silence H2AX.
Results: 1. Successfully constructed the ECA109 cell line with silence H2AX gene. 2. Ionizing radiation could cause r-H2AX and MCPH1 protein expression increase, as the same as nuclear focus increase of r- H2AX and MCPH1. 3. The protein level and nucleus focus of r-H2AX and MCPH1 are significantly reduced in ECA109 after silence H2AX.
Conclusion: MCPH1 is the part of DNA damage response triggered by ionizing radiation and is located in the damage response downstream and can be regulated by H2AX.
Keywords
Esophageal cancer, Ionizing radiation, DNA double-strand breaks, γ-H2AX, MCPH1.
Introduction
MCPH1 plays an important role in DNA damage response, especially participating in cell-cycle checkpoint control [1]. Previous literature has indicated that phosphorylation of H2AX was the early critical step in DNA damage repair pathways [2-4], but the relationship of MCPH1 and H2AX is not clear. Here we aimed to study the role of MCPH1 in DNA damage response and reveal the relationship between MCPH1 and H2AX.
Materials and Methods
Cell culture
Esophageal cancer cell line ECA109 was bought from Genechem (Shanghai). Cells were cultured in RPMI-1640 medium plus 10% FBS, 100 U/ml penicillin and 100 μg/ml streptomycin in a glass flask. Cells were maintained in humidified 5% CO2/95% air at 37ºC and subcultured every 2 to 3 d.
H2AX silencing
Lentivirus-mediated H2AX RNAi vector containing shRNA sequence was from Genechem. After transfection with viral vector or control vector, 5 μg/ml puromycin was added to the medium for 72 h for cell selection. Then transfection efficiency was observed by fluorescence microscopy. Cells were subcultured, and RNA and protein were extracted at the 1st, 5th, and 10th generation, respectively. Then RT-PCR and Western blot were used to measure the H2AX silencing efficiency. The primers are as follows: upstream primers: 5' CAGTGCTGGAGTACCTCAC 3'; downstream primers: 5' CTGGATGTTGGGCAGGAC 3'.
Ionizing radiation
6MV-X Siemens linear accelerator at a dose rate of 2 Gy/min was used. Compensation film was put at 5 cm underneath the flask, and cells with or without H2AX silencing were exposed to 8 Gy at a field of 20 × 20 cm size and 100 cm source-skin distance at room temperature. After 1 hour of radiation, total protein was extracted and stored at -20°C. Coomassie brilliant blue assay was used for quantification of proteins.
Western blot
Sample preparation: 20 μg protein from ECA109 cells after radiation was mixed with 4X loading buffer at 3:1. Then samples were centrifuged, boiled at 100°C for 5 min, and cooled down for use.
Sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE
20 μg protein samples were separated by 12% or 10% separating gel and wet-transferred to a PVDF membrane. After being blocked with 5% fat-free milk in TBST for 1 hour at 37°C, the membrane was incubated with primary antibody (anti-γ-H2AX 1:1000, anti-MCPH1 1:200, anti-GAPDH 1:500) at 4°C overnight. Then the membrane was washed 4 times for 10 min each in TBST buffer and incubated with fluorescence labeled secondary antibody at 37°C for 1 h, avoiding exposure to light. After 4 times’ washing for 10 min each in TBST buffer, the membrane was scanned by infrared imaging system. The protein expression level was measured by quantification of Optical Density (OD) of each band.
Immunofluorescence
Cells kept in log phase were digested with trypsin and resuspended at 1 × 106/ml, and then 100 μL cell suspension was seeded into a new dish, with another 2 ml of fresh medium. Cells were maintained in an incubator at 37°C for 24 h and exposed to the ionizing radiation for 1 h. After washing 3 times with PBS, cells were fixed with 4% PFA for 15 min. Then the fixative was discarded and cells were washed with PBS 3 times for 10 min each with gentle shaking. Cells were permeabilized with 0.5% Triton X-100 3 times for 5 min each and washed in PBS 3 times for 10 min each, again with gentle shaking. Then the samples were blocked with goat serum for 1 hour and further incubated with primary antibody (anti-γ- H2AX 1:100, anti-MCPH1 1:100) at 37°C for 2 h. After washing 3 times in PBS with gentle shaking, cells were exposed to Cy-3 labeled goat anti-rabbit secondary antibody (1:100) containing DAPI (1:2000) for 2 h at 37°C in a wet box. Then the unbound secondary antibody was removed by washing 3 times for 10 min each with PBS with gentle shaking, and slides were mounted with mounting medium and visualized in a Nikon ECLIPSE E800 microscope.
Co-immunoprecipitation
ECA109 cells with or without H2AX silencing were exposed to 8 Gy for 1 h, and total protein was extracted. 80 μg protein was incubated with 3 μL anti-γ-H2AX, anti-MCPH1, anti- H2AX rabbit antibody or rabbit IgG negative control, 2.5 μL PMSF, and enough IPH to bring the total volume to 250 μL. After 1 hour of incubation at 4°C, 20 μL protein A beads were added into the protein-antibody reaction system and incubated overnight at 4°C. After centrifugation at 12000 rpm for 3 min at 4°C, supernatants were discarded and 400 μL IPH was added into the IP reaction system; the tube then was rotated for 20 min at 4°C. This washing step was repeated 4 times. Then centrifugation was performed at 12000 rpm for 3 min at 4°C, and supernatants were discarded as much as possible. 10-15 μL 2X buffer was added into the beads and mixed gently. Then the beads were boiled for 5 min and centrifuged at 12000 rpm for 1 min at room temperature. The supernatant was subjected to Western blot, and expression levels of γ-H2AX, MCPH1, and H2AX were detected.
Statistical analysis
The statistical analysis was performed using SPSS17.0 software. The results were shown as mean ± SD (x̄ ± SD). One-way ANOVA was used to compare the difference between groups.
Results
Esophageal cancer cell line ECA109 with stable lowlevel expression of H2AX was established by knockdown of H2AX
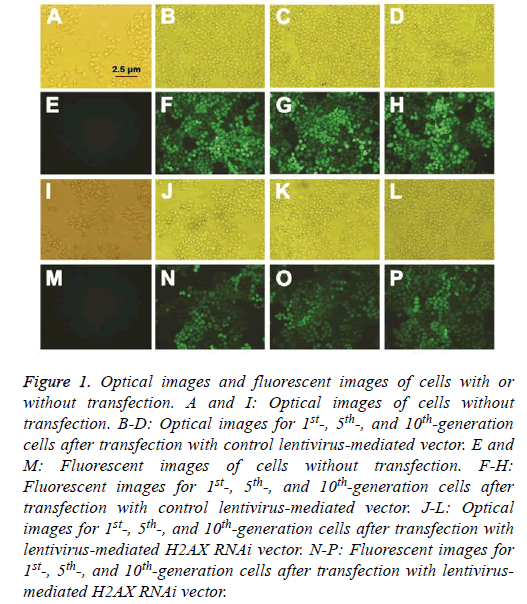
Lentivirus-mediated vectors were double-labeled with fluorescence and antibiotic. The lentivirus-mediated H2AX RNAi vector and control vector were transfected into ECA109 cells, and an antibiotic was used for cell selection. The transfection efficiency was observed by fluorescence microscope. As shown in Figure 1, the transfection efficiency was up to 99%.
Figure 1: Optical images and fluorescent images of cells with or without transfection. A and I: Optical images of cells without transfection. B-D: Optical images for 1st-, 5th-, and 10th-generation cells after transfection with control lentivirus-mediated vector. E and M: Fluorescent images of cells without transfection. F-H: Fluorescent images for 1st-, 5th-, and 10th-generation cells after transfection with control lentivirus-mediated vector. J-L: Optical images for 1st-, 5th-, and 10th-generation cells after transfection with lentivirus-mediated H2AX RNAi vector. N-P: Fluorescent images for 1st-, 5th-, and 10th-generation cells after transfection with lentivirusmediated H2AX RNAi vector.
H2AX silencing effect was detected at both mRNA and protein expression level
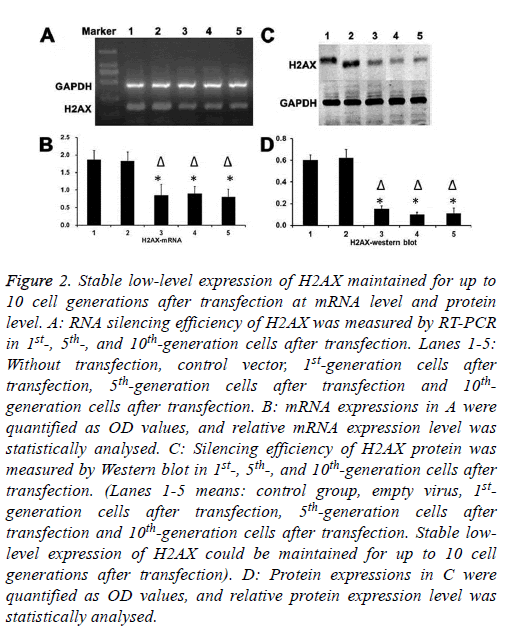
As shown in Figure 2, stable low-level expression of H2AX could be maintained for up to 10 cell generations after transfection.
Figure 2: Stable low-level expression of H2AX maintained for up to 10 cell generations after transfection at mRNA level and protein level. A: RNA silencing efficiency of H2AX was measured by RT-PCR in 1st-, 5th-, and 10th-generation cells after transfection. Lanes 1-5: Without transfection, control vector, 1st-generation cells after transfection, 5th-generation cells after transfection and 10thgeneration cells after transfection. B: mRNA expressions in A were quantified as OD values, and relative mRNA expression level was statistically analysed. C: Silencing efficiency of H2AX protein was measured by Western blot in 1st-, 5th-, and 10th-generation cells after transfection. (Lanes 1-5 means: control group, empty virus, 1stgeneration cells after transfection, 5th-generation cells after transfection and 10th-generation cells after transfection. Stable lowlevel expression of H2AX could be maintained for up to 10 cell generations after transfection). D: Protein expressions in C were quantified as OD values, and relative protein expression level was statistically analysed.
Ionizing radiation could increase both the γ-H2AX expression level and the number of its nuclear foci
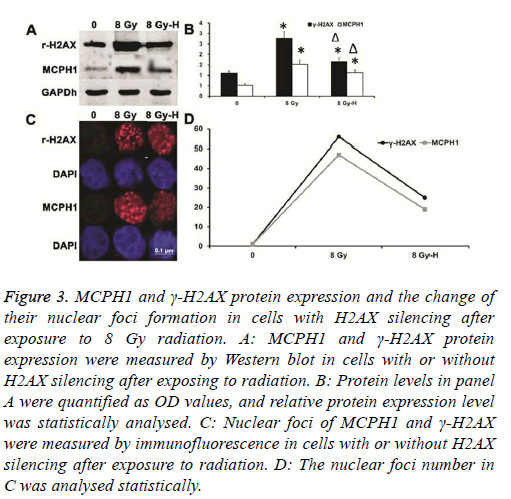
Also, the ionizing radiation showed the same effect as MCPH1. When silencing H2AX expression, not only was the up-regulation of γ-H2AX expression inhibited, but the increase of γ-H2AX nuclear foci number was also weakened after ionizing radiation. In the meantime, up-regulation of MCPH1 induced by ionizing radiation was suppressed, and the increase of its nuclear foci number was inhibited (Figure 3).
Figure 3: MCPH1 and γ-H2AX protein expression and the change of their nuclear foci formation in cells with H2AX silencing after exposure to 8 Gy radiation. A: MCPH1 and γ-H2AX protein expression were measured by Western blot in cells with or without H2AX silencing after exposing to radiation. B: Protein levels in panel A were quantified as OD values, and relative protein expression level was statistically analysed. C: Nuclear foci of MCPH1 and γ-H2AX were measured by immunofluorescence in cells with or without H2AX silencing after exposure to radiation. D: The nuclear foci number in C was analysed statistically.
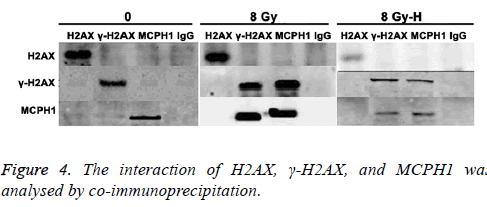
Interaction of H2AX, γ-H2AX and MCPH1 was analysed by co-immunoprecipitation
As shown in Figure 4, when cells were not exposed to radiation, there was no interaction between H2AX, γ-H2AX, and MCPH1. After ionizing radiation, the MCPH1 interacted with γ-H2AX but not H2AX. Moreover, this interaction was attenuated after silencing H2AX.
Discussion
The number of new esophageal cancer cases was 482300 worldwide, and 406800 esophageal cancer deaths occurred in 2010 [5], ranked the sixth in cancer deaths. In China, incidence of esophageal cancer accounts for 50% of the total cases worldwide. In Hebei province, its incidence is as high as 100/100000. Even though therapy techniques-such as surgery, radiotherapy, chemotherapy, and targeted therapy-are developing fast, the 5 y survival rate of early esophageal cancer after surgery remains only 30%. Most of the failed cases are due to cancer recurrence [6-8].
Early-stage esophageal cancer may be totally asymptomatic or may present with mild nonspecific symptoms; thus, most patients went to the hospital when the esophageal cancer had progressed rapidly. Radiotherapy becomes the major treatment for middle/late-stage esophageal cancer; however, the recurrence of the tumor is still the main cause of treatment failure. Therefore, radiotherapists continue to focus on improving the efficacy of radiotherapy for esophageal cancer. Improvement in fundamental research may help reveal details of the mechanism of the radiotherapy. Increasing the sensitivity of radiotherapy through optimizing conditions is the key aspect of radiologists’ research.
MCPH1 is involved in DNA damage response, especially in the cell-cycle checkpoint. However, the process through which the MCPH1 regulates DNA damage response has not been clear. MDC1, BRCA1, and 53BP1 are the well-known regulatory proteins in DNA damage response pathways. The newly discovered regulatory protein MCPH1 was found to enhance the activation and transduction of DNA damage response. Wood et al. found that MCPH1 foci formed in nuclei after HeLa cell exposure to ionizing radiation. Not only radiation, but all stimulation leading to DNA damage, could induce these foci. MCPH1 foci were co-localized with γ- H2AX in the nuclei, indicating that MCPH1 could be re localized to the position where DNA damage occurred. The same observation was also found in esophageal cancer cells after radiotherapy. Phosphorylation of H2AX is critical for MCPH1 foci formation after DNA damage.
The consequences when MCPH1 was knocked out with RNAi have been described in some reports [9-12]. In our study, there was no interaction between MCPH1 and phosphorylated H2AX (γ-H2AX) without ionizing radiation treatment, while after radiation, MCPH1 interacted closely with γ-H2AX. When silencing H2AX, the interaction weakened. Moreover, both the γ-H2AX foci formation in nuclei and the up-regulation of its expression induced by ionizing radiation decreased when silencing H2AX. Accompanying it, the number of MCPH1 foci decreased and protein expression declined. All these results suggested that MCPH1 was located in the damage response downstream and regulated by H2AX.
MCPH1 is very important to brain development and tumor suppression. One of the recent studies showed that MCPH1 was not heterozygous and was abnormally expressed in breast cancer [11]. More research on MCPH1 knockout mice will help in identifying whether MCPH1 is a tumor suppressor. If MCPH1 could be gradually applied to clinical trials as a tumor suppressor in the future, it may provide relief for patients and help reduce the incidence of the disease. Thus, as a potential tumor suppressor, there is great significance in continuing research on MCPH1.
Acknowledgements
This study was supported by Province Natural Science Foundation of Hebei (H201501143) and Hebei University Medical Discipline special fund construction projects (2014B1001) and Key Research Fund in Affiliated Hospital of Hebei University (2015Z001) and the key laboratory project of tumor radiotherapy and chemotherapy mechanism and regulation in Hebei province.
Conflicts of Interest
The authors declare that they have no conflict of interest.
References
- Wood JL, Singh N, Mer G, Chen J. MCPH1 functions in an H2AX-dependent but MDC1-independent pathway in response to DNA damage. J Biol Chem 2007; 282: 35416-35423.
- Wu CY, Kang HY, Yang WL. Critical role of monoubiquitination of histone H2AX protein in histone H2AX phosphorylation and DNA damage response. J Biol Chem 2011; 286: 30806-30815.
- Jemal A, Bray F, Center MM, Ferlay J, Ward E, Forman D. Global cancer statistics. CA Cancer J Clin 2011; 61: 69-90.
- Boonstra JJ, Kok TC, Wijnhoven BP. Chemotherapy followed by surgery versus surgery alone in patients with resectable oesophageal squamous cell carcinoma: long-term results of a randomized controlled trial. BMC Cancer 2011; 11: 181.
- Yamashita H, Okuma K, Wakui R, Kobayashi-Shibata S, Ohtomo K, Nakagawa K. Details of recurrence sites after elective nodal irradiation (ENI) using 3D-conformal radiotherapy (3D-CRT) combined with chemotherapy for thoracic esophageal squamous cell carcinoma-a retrospective analysis. Radiother Oncol 2011; 98: 255-260.
- Ono S, Fujishiro M, Niimi K. Long-term outcomes of endoscopic submucosal dissection for superficial esophageal squamous cell neoplasms. Gastrointest Endosc 2009; 70: 860-866.
- Xu X, Lee J, Stern DF. Microcephalin is a DNA damage response protein involved in regulation of CHK1 and BRCA1. J Biol Chem 2004; 279: 34091-34094.
- Lin SY, Rai R, Li K, Xu ZX, Elledge SJ. BRIT1/MCPH1 is a DNA damage responsive protein that regulates the Brca1-Chk1 pathway, implicating checkpoint dysfunction in microcephaly. Proc Natl Acad Sci USA 2005; 102: 15105-15109.
- Rai R, Dai H, Multani AS. BRIT1 regulates early DNA damage response, chromosomal integrity, and cancer. Cancer Cell 2006; 10: 145-157.
- Alderton GK, Galbiati L, Griffith E. Regulation of mitotic entry by microcephalin and its overlap with ATR signalling. Nat Cell Biol 2006; 8: 725-733.