Research Article - Biomedical Research (2017) Volume 28, Issue 2
Epidemiological features and genetic variation of human respiratory syncytial virus (HRSV) infection in Chungnam, Korea
Kwisung Park1#, Donguk Kim1,2#, Jeongmin Seong3, Inchul Shin1, Jayoung Hong1, Seongmin Park1,Shien-Young Kang2, Hyekyung Lee4*1Chungcheongnam-Do Health and Environment Research Institute, Daejeon, Korea
2College of Veterinary Medicine, Chungbuk National University, Cheongju, Korea
3Department of Dental Hygiene, College of Health Science, Kangwon National University, Samcheok, Korea
4Department of Nursing, College of Nursing and Health, Kongju National University, Gonju, Korea
#These authors contributed equally to this work
- *Corresponding Author:
- Hyekyung Lee
Department of Nursing, College of Nursing and Health
Kongju National University, Republic of Korea
Accepted on July 26, 2016
Abstract
We investigated the epidemiological patterns and genetic variation of Human Respiratory Syncytial Virus (HRSV) isolated from Chungnam, Korea, during 2011-2014. It was screened 3,531 nasopharyngeal aspirate samples from patients with acute respiratory symptoms for HRSV and other respiratory viruses by real-time reverse-transcription polymerase chain reaction. Temporal distribution of the HRSV epidemic showed seasonal prevalence for early winter and mainly founded in children’s less than 3 years of age. Ninety-two (29.8%) HRSV-positive patients were co-infected with other respiratory viruses including human rhinovirus, adenovirus, and human bocavirus. In phylogenetic analysis of the isolated strains based on G-protein gene sequences, only one isolate (CNA013- 11/07-2011/Korea) was the GA5 genotype, 17 were the NA1 genotype, and all strains isolated after 2012 belonged to the ON1 genotype among the 47 HRSV-A strains. All 18 HRSV-B strains isolated in this study belonged to the BA4 genotype. Continued studies on HRSV epidemiology and genetic diversity of circulating HRSV isolates are necessary to facilitate the development of efficacious vaccines and antiviral agents.
Keywords
HRSV, RT-PCR, G-protein, Phylogenetic tree.
Introduction
Human Respiratory Syncytial Virus (HRSV) is the second most common cause of acute respiratory infection in infants and young children, and is found mainly from late fall to spring in temperate countries [1,2]. Most infants experience HRSV infection in the first year of life, and nearly all are infected by 2 years of age [3]. HRSV, a member of the Pneumovirus genus of the Paramyxoviridae family, was first identified in 1956 [4]. The enveloped HRSV has a negative-sense, non-segmented single-stranded RNA genome that encodes for 11 distinct proteins, including Fusion Protein (F-protein) and attachment (G-protein) Glycoproteins that are capable of binding host cell receptors and are often targeted by the body’s antibody responses [5]. HRSV strains are classed into 2 major genogroups (A and B) based on the G-protein gene sequence, which is the main site of diversifying selection in the genome. As such, molecular analysis of the G-protein gene has been used to determine the HRSV genotype and to characterize diversity in HRSV strains [6,7]. Based on this method, HRSVA is divided into 11 genotypes (e.g., GA1-7, SAA1, NA1-2, and ON1) and HRSV-B into 23 genotypes (e.g., GB1-4, SAB1-4, URU1-2, BA1-12, and THB). Various viral genotypes co-circulate in epidemic seasons, but a new genotype can replace previously dominant strains [1,8]. Since no HRSV vaccine is currently available, the on-going regional surveillance and molecular characterization of circulating HRSV strains are necessary for vaccine and antiviral agent development [9]. In this study, we report an overview of the epidemiology and genetic diversity of HRSV isolates circulating within Chungnam, Korea, during a 4-year period from 2011 to 2014.
Materials and Methods
Sample collection
The samples used in this study were collected under the guidelines of the Korea Influenza and Respiratory Viruses Surveillance System with written informed consent. Between January 2011 and December 2014, a total of 3,531 nasopharyngeal specimens were obtained from patients presenting with symptoms of acute respiratory tract disease admitted to the local hospital in Chungnam province, Korea. Nasopharyngeal swabs were collected in 3 mL of universal viral transport medium (BD, Maryland, USA) and stored at -70ºC prior to analysis.
Real-time reverse transcription (RT) polymerase chain reaction (PCR)
Viral RNA was extracted from nasopharyngeal specimens using the Maxwell 16 Viral Total Nucleic Acid Purification kit (Promega, Madison, USA) per the manufacturer’s instructions. Viral RNA was eluted in 50 μL of elution buffer and stored at -70ºC. Samples were screened for the presence of 14 common viral respiratory pathogens with the following manufacturer’s instructions by commercial real-time PCR kit (Kogene Biotech; Seoul, Korea); Adenovirus (ADV), Human Bocavirus (HBoV), Parainfluenza Virus (PIV) Type 1/2/3, Human Metapneumovirus (HMPV), Human Coronavirus (HCoV) 229E/OC43/NL63, Human Rhinovirus (HRV), influenza A/B and HRSV A/B.
RT-PCR and sequencing
HRSV-A and B positive RNA samples were subjected to Gprotein gene amplification by RT-PCR using the Access RTPCR system (Promega, Madison, USA) per the manufacturer’s instructions with the following published primer sets: ABG490 located in the G-protein region (5'- ATGATTWYCAYTTTGAAGTGTTC-3') and F164 located in F-protein region (5'-GTTATGACACTGGTATACCAACC-3') [10]. RT-PCR was conducted under the following conditions: RT at 45ºC for 45 min and PCR at 95ºC for 5 min, 35 cycles (94ºC for 45 sec, 50ºC for 45 sec and 72ºC for 1 min) and 72ºC for 7 min.
PCR products were purified using the QIA quick PCR purification kit (Qiagen, Germany). Purified DNA was added in a reaction mixture containing 2 μL of Big Dye terminator reaction mix (ABI Prism Big Dye Terminator Cycle Sequencing Kit; Perkin-Elmer Applied Bio systems, USA) and 2 pmol of sense or antisense primer. Sequencing reactions were subjected to initial denaturation at 94ºC for 1 min and 25 cycles consisting of 94ºC for 10 sec, 50ºC for 5 sec, and 60ºC for 4 min in a Gene Amp PCR system 2700 (Applied Bio systems). PCR products were purified by precipitation in 100% cold ethanol and 3 M sodium-acetate (pH 5.8) and then loaded on an automated 3100 Genetic Analyser (Applied Bio systems).
Phylogenetic analysis
G-protein gene sequences for 47 HRSV-A isolates and 18 HRSV-B isolates were compared with the reference sequences using Clustal W version. 2.1 [11]. Phylogenetic relationships among the isolates and reference strains were determined using MEGA software version. 6.0. Maximum composite likelihood was used as the substitution method, followed by the neighbour-joining method to reconstruct the phylogenetic tree [12]. The reliability of the constructed phylogenetic tree was determined by bootstrap re-sampling of 1,000 replicates.
Nucleotide sequence accession numbers
The HRSV candidate sequences reported here were deposited in the GenBank sequence database: accession numbers KU681134-KU681172, KU245519-KU245529, and KU724059-KU724073.
Results
Epidemiological features of HRSV
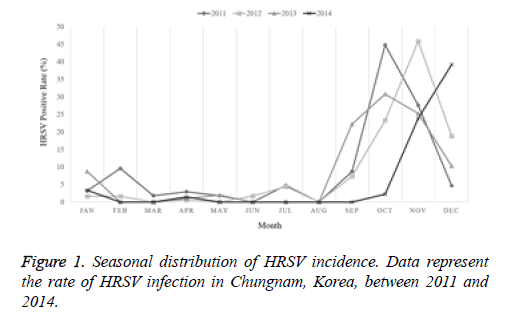
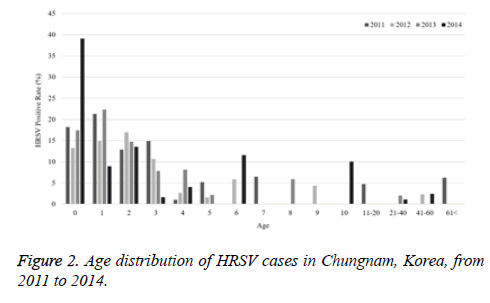
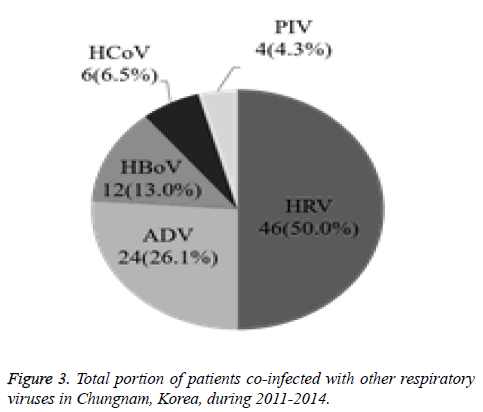
The current study investigated the epidemic patterns and genetic diversity of HRSV isolates circulating within Chungnam, Korea, from 2011 to 2014. For this, 3,531 samples were obtained from patients presenting with acute respiratory symptoms, under the guidelines of the Korea Influenza and Respiratory Viruses Surveillance System. HRSV-positive isolates were subjected to G-protein gene sequencing and analysis. A total of 309 HRSV strains were identified: 92 isolates in 2011, 84 isolates in 2012, 91 isolates in 2013, and 42 isolates in 2014 as shown in Table 1. In this period, the annual peak of HRSV infection occurred during the early winter season, with incidences peaking from September to December (16.8%-31.9%) and high incidence being noted in November (31.9%) as shown in Figure 1. The majority of patients infected with HRSV were children under 3 years of age, accounting for 89.3% of all cases: 9.1% for infants aged 0 year, 36.6% for infants aged 1 year, 30.4% for children aged 2 years, and 13.2% for children aged 3 years as shown in Figure 2. The male-to-female ratio of infected individuals was 1.27:1. Co-infection with other respiratory viruses was detected in 92 patients (29.8%), and included HRV (46 samples), ADV (24 samples), HBoV (12 samples), HCoV (6 samples), and PIV (4 samples) as shown in Figure 3.
| Age | No. of samples (Positive number) | |||||
|---|---|---|---|---|---|---|
| 2011 | 2012 | 2013 | 2014 | Total | ||
| No. | Rate (%) | |||||
| 0 | 11 (2) | 68 (9) | 46 (8) | 23 (9) | 148 (28) | 18.92 |
| 1 | 188 (40) | 180 (27) | 159 (35) | 123 (11) | 650 (113) | 17.38 |
| 2 | 155 (20) | 183 (31) | 204 (30) | 96 (13) | 638 (94) | 14.73 |
| 3 | 134 (20) | 94 (10) | 128 (10) | 61 (1) | 417 (41) | 9.83 |
| 4 | 98 (1) | 76 (2) | 62 (5) | 25 (1) | 261 (9) | 3.45 |
| 5 | 58 (3) | 63 (1) | 46 (1) | 25 (0) | 192 (5) | 2.6 |
| 6 | 37 (0) | 34 (2) | 45 (0) | 26 (3) | 142 (5) | 3.52 |
| 7 | 31 (2) | 28 (0) | 28 (0) | 10 (0) | 97 (2) | 2.06 |
| 8 | 33 (0) | 24 (0) | 17 (1) | 9 (0) | 83 (1) | 1.2 |
| 9 | 16 (0) | 23 (1) | 21 (0) | 14 (0) | 74 (1) | 1.35 |
| 10 | 15 (0) | 14 (0) | 20 (0) | 10 (1) | 59 (1) | 1.69 |
| Nov-20 | 42 (2) | 64 (0) | 42 (0) | 69 (0) | 217 (2) | 0.92 |
| 21-40 | 43 (0) | 57 (0) | 50 (1) | 91 (1) | 241 (2) | 0.83 |
| 41-60 | 34 (0) | 44 (1) | 42 (0) | 82 (2) | 202 (3) | 1.49 |
| >60 | 32 (2) | 44 (0) | 23 (0) | 11 (0) | 110 (2) | 1.82 |
| Total | 927 (92) | 996 (84) | 933 (91) | 675 (42) | 3531 (309) | 8.75 |
Table 1. The number of RSV detected in Chungnam Korea by age distribution from 2011 to 2014.
Phylogenetic analysis of HRSV-A and B
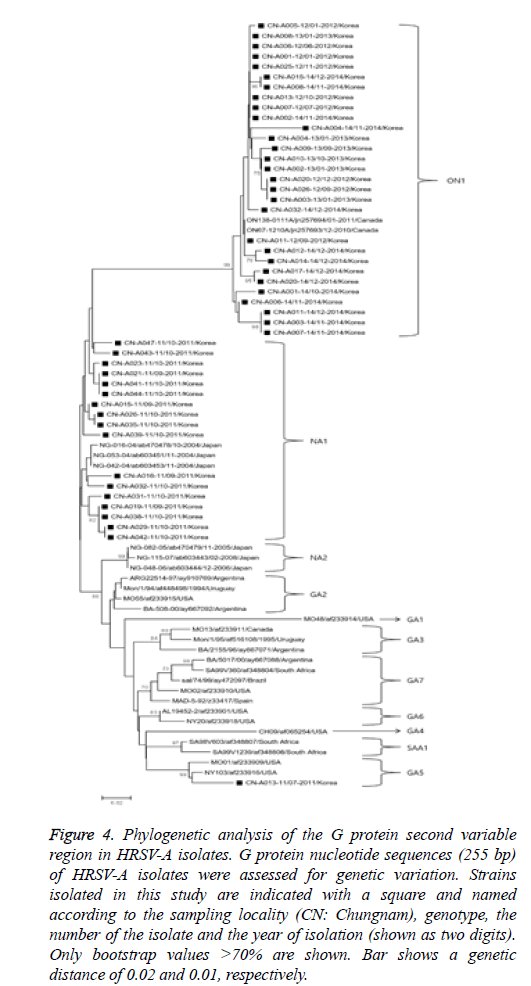
Sixty-five partial G-protein nucleotide sequences from isolates obtained in this study were used to construct phylogenetic trees (HRSV-A and -B) with reference strains deposited in the GenBank database as shown in Figure 4. In the HRSV-A phylogenetic analysis, 255 bp G-protein nucleotide sequences from 47 randomly selected strains were used to construct a phylogenetic tree with 30 references of the same type obtained from GenBank database. The HRSV-A strains were categorized into 11 distinct genetic groups, which were supported by high bootstrap values and previously reported clusters, including GA1-7 [3,13], NA1-2 [1], SAA1 [14], and ON1 [15]. Specifically, the 47 HRSV-A strains isolated in our study belonged to three previously defined clusters, GA5 (1 isolate), NA1 (17 isolates), and ON1 genotypes (29 isolates). Of note, strains isolated in 2011 showed an 8.8-12.2% and 10.1-18.8% nucleotide divergence from the GA5 and ON1 genotypes, respectively.
Figure 4: Phylogenetic analysis of the G protein second variable region in HRSV-A isolates. G protein nucleotide sequences (255 bp) of HRSV-A isolates were assessed for genetic variation. Strains isolated in this study are indicated with a square and named according to the sampling locality (CN: Chungnam), genotype, the number of the isolate and the year of isolation (shown as two digits). Only bootstrap values >70% are shown. Bar shows a genetic distance of 0.02 and 0.01, respectively.
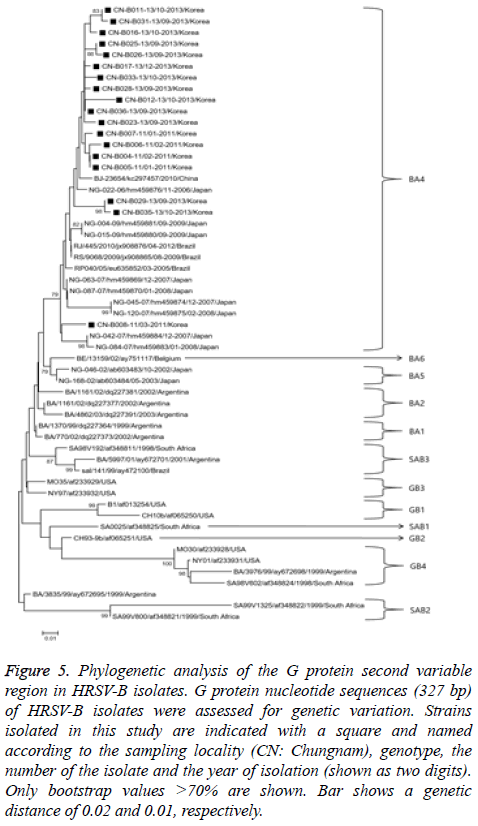
Figure 5: Phylogenetic analysis of the G protein second variable region in HRSV-B isolates. G protein nucleotide sequences (327 bp) of HRSV-B isolates were assessed for genetic variation. Strains isolated in this study are indicated with a square and named according to the sampling locality (CN: Chungnam), genotype, the number of the isolate and the year of isolation (shown as two digits). Only bootstrap values >70% are shown. Bar shows a genetic distance of 0.02 and 0.01, respectively.
The HRSV-B phylogenetic analysis used 327 bp G-protein nucleotide sequences from 18 randomly selected HRSV-B to construct a phylogenetic tree with sequences from 38 reference strains obtained from the GenBank database as shown in Figure 5. Strains were categorized into 12 distinct genetic groups, which were supported by high bootstrap values and previously reported clusters, including BA1-6 [16], GB1-4 [13], and SAB1-3 [14]. All 18 HRSV-B strains isolated in our study belonged to BA-4 genotype.
Discussion
HRSV is a leading cause of acute respiratory syndrome among children and immunocompromised adults worldwide [17,18].Of patients presenting with acute respiratory symptoms, 8.75% were HRSV-positive, the majority of which were less than 3 years of age in this study. Similarly, HRSV infection incidence was reported to be 8.9% among hospitalized patients in Thailand [19], with an increased incidence in children less than 5 years of age [9]. Several previous studies demonstrate that HRSV-associated acute respiratory syndrome occurs mainly from late fall to winter season [9,20,21], consistent with our data. Moreover, 92 HRSV-positive patients were co-infected with additional respiratory viruses, including HRV, ADV, HBoV, HCoV, and PIV’s. Advances in diagnostic methods have increased the cases of co-detection, and its clinical significance should be interpreted cautiously [22]. Phylogenetic analysis of 47 selected HRSV-A isolates revealed their associations with the previously defined genotypes GA5, NA1, and ON1. The HRSV GA5 genotype include strains isolated in Europe, Uruguay, and USA in early 1990’s [23,24], and only one Korean isolate from our study showed phylogenetic similarity to this genotype (CNA013- 11/07-2011/Korea). NA1 genotype strains diverged from the GA2 genotype, leading to widespread HRSV-A outbreaks in Niigata, Japan, since 2006 [1] and in Chungnam, Korea, in 2011. Analysis of HRSV-A strains isolated in Ontario, Canada during winter 2010-2011 delineated separation of the ON1 genotype from NA1 as a result of a 72-nucleotide duplication in the G-protein C-terminus [15], and this genotype predominated in Chungnam from 2012 to 2014. HRSV may evolve with a distinct pattern determined by viral strain, host, and local characteristics. Particularly, HRSV-A strains with genetic replacement of the G protein were isolated in the same region through successive epidemics [25].
All 18 HRSV-B strains isolated in our study belonged to the BA4 genotype, which harbours 60-nucleotide duplication in the G-protein gene [26]. The BA1-6 genotypes were classified in genetic analysis of G-protein in HRSV-B isolates collected in Buenos Aires, Argentina, in 1999 [26]. BA4 strains have circulated worldwide since 2001 and display a high mutation rate [27,28]; thus, the rapid worldwide spread of the BA genotype may indicate a selective advantage over other circulating strains [29]. There were distinct lineages of the G protein among both subgroups A and B between geographic regions, and it suggested the positive immune selection in the same region [25].
Recently, it was reported for a vaccine study using HRSV G protein fragment in order to represent a candidate immunogen for HRSV vaccine [30]. The analysis of HRSV epidemiology and G-protein gene variability in the isolates from Chungnam, Korea revealed the patterns of genetic variation, and therefore it is believed that this research may be useful in the future development of vaccines and anti-viral therapies against HRSV infection.
Acknowledgements
This study was funded and technically supported by KCDC (Korea Centers for Disease Control and Prevention) and Kongju University.
References
- Shobugawa Y, Saito R, Sano Y, Zaraket H, Suzuki Y, Kumaki A, Dapat I, Oguma T, Yamaguchi M, Suzuki H. Emerging genotypes of human respiratory syncytial virus subgroup A among patients in Japan. J ClinMicrobiol 2009; 47: 2475-2482.
- Borchers AT, Chang C, Gershwin ME, Gershwin LJ. Respiratory syncytial virus-a comprehensive review. Clin Rev Allergy Immunol 2013; 45: 331-379.
- Cane PA. Molecular epidemiology of respiratory syncytial virus. Rev Med Virol 2001; 11: 103-116.
- Collins PL, Crowe JE Jr. Respiratory syncytial virus and Metapneumovirus. Fields Virol Lippincott Williams Wilkins PA (5th edn.) 2007; 1601-1646.
- Sullender WM. Respiratory syncytial virus genetic and antigenic diversity. ClinMicrobiol Rev 2000; 13: 1-15.
- Peret TC, Hall CB, Schnabel KC, Golub JA, Anderson LJ. Circulation patterns of genetically distinct group A and B strains of human respiratory syncytial virus in a community. J Gen Virol 1998; 79: 2221-2229.
- Peret TC, Hall CB, Hammond GW, Piedra PA, Storch GA, Sullender WM, Tsou C, Anderson LJ. Circulation patterns of group A and B human respiratory syncytial virus genotypes in 5 communities in North America. J Infect Dis 2000; 181: 1891-1896.
- Rebuffo-Scheer C, Bose M, He J, Khaja S, Ulatowski M, Beck ET, Fan J, Kumar S, Nelson MI, Henrickson KJ. Whole genome sequencing and evolutionary analysis of human respiratory syncytial virus A and B from Milwaukee, WI 1998-2010. PLoS One 2011; 6: 25468.
- Arnott A, Vong S, Mardy S, Chu S, Naughtin M, Sovann L, Buecher C, Beaute J, Rith S, Borand L, Asgari N, Frutos R, Guillard B, Touch S, Deubel V, Buchy P. A study of the genetic variability of human respiratory syncytial virus (HRSV) in Cambodia reveals the existence of a new HRSV group B genotype. J ClinMicrobiol 2011; 49: 3504-3513.
- Parveen S, Sullender WM, Fowler K, Lefkowitz EJ, Kapoor SK. Genetic variability in the G protein gene of group A and B respiratory syncytial viruses from India. J ClinMicrobiol 2006; 44: 3055-3064.
- Thompson JD, Higgins DG, Gibson TJ. Clustal W-improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res 1994; 22: 4673-4680.
- Tamura K, Stecher G, Peterson D, Filipski A, Kumar S. MEGA6: Molecular Evolutionary Genetics Analysis version 6.0. MolBiolEvol 2013; 30: 2725-2729.
- Galiano MC, Palomo C, Videla CM, Arbiza J, Melero JA, Carballal G. Genetic and antigenic variability of human respiratory syncytial virus (groups a and b) isolated over seven consecutive seasons in Argentina. J ClinMicrobiol 2005; 43: 2266-2273.
- Venter M, Madhi SA, Tiemessen CT, Schoub BD. Genetic diversity and molecular epidemiology of respiratory syncytial virus over four consecutive seasons in South Africa: identification of new subgroup A and B genotypes. J Gen Virol 2001; 82: 2117-2124.
- Eshaghi A, Duvvuri VR, Lai R, Nadarajah JT, Li A, Patel SN, Low DE, Gubbay JB. Genetic variability of human respiratory syncytial virus a strains circulating in Ontario-a novel genotype with 72 nucleotide gene duplication. PLoS One 2012; 7: 32807.
- Trento A, Casas I, Calderon A, Garcia-Garcia ML, Calvo C, Perez-Brena P, Melero JA. Ten years of global evolution of the human respiratory syncytial virus BA genotype with 60-nucleotide duplication in the G protein gene. J Virol 2010; 84: 7500-7012.
- Johnson PR, Spriggs MK, Olmsted RA, Collins PL. The G glycoprotein of human respiratory syncytial viruses of subgroups A and B: extensive sequence divergence between antigenically related proteins. ProcNatlAcadSci USA 1987; 84: 5625-5629.
- Nair H, Simoes EA, Rudan I, Gessner BD, Azziz-Baumgartner E, Zhang JS, Feikin DR, Mackenzie GA, Moisi JC, Roca A, Baggett HC, Zaman SM, Singleton RJ, Lucero MG, Chandran A, Gentile A, Cohen C, Krishnan A, Bhutta ZA, Arguedas A, Clara AW, Andrade AL, Ope M, Ruvinsky RO, Hortal M, McCracken JP, Madhi SA, Bruce N, Qazi SA, Morris SS, El Arifeen S, Weber MW, Scott JA, Brooks WA, Breiman RF, Campbell H. Severe Acute Lower Respiratory Infections Working Group. Global and regional burden of hospital admissions for severe acute lower respiratory infections in young children in 2010-a systematic analysis. Lancet 2013; 381: 1380-1390.
- Fry AM, Chittaganpitch M, Baggett HC, Peret TC, Dare RK, Sawatwong P, Thamthitiwat S, Areerat P, Sanasuttipun W, Fischer J, Maloney SA, Erdman DD, Olsen SJ. The burden of hospitalized lower respiratory tract infection due to respiratory syncytial virus in rural Thailand. PLoS One 2010; 5: 15098.
- Law BJ, Carbonell-Estrany X, Simoes EA. An update on respiratory syncytial virus epidemiology: a developed country perspective. Respir Med 2002; 96: 1-7.
- Kim CK, Choi J, Callaway Z, Kim HB, Chung JY. Clinical and epidemiological comparison of human metapneumovirus and respiratory syncytial virus in seoul, Korea. J Korean Med Sci 2010; 25: 342-347.
- Roh EJ, Chang YP, Kim JK, Rheem IS, Park KS, Chung EH. Clinical significance of codetection of the causative agents for acute respiratory tract infection in hospitalized children. Korean J Pediatr 2009; 52: 661-666.
- Cane PA, Pringle CR. Respiratory syncytial virus heterogeneity during an epidemic: analysis by limited nucleotide sequencing (SH gene) and restriction mapping (N gene). J Gen Virol 1991; 72: 349-357.
- Garcia O, Martin M, Dopazo J, Arbiza J, Frabasile S, Russi J, Hortal M, Perez-Brena P, Martinez I, Garcia-Barreno B, Meleroi JA. Evolutionary pattern of human respiratory syncytial virus (subgroup A)-cocirculating lineages and correlation of genetic and antigenic changes in the G glycoprotein. J Virol 1994; 68: 5448-5459.
- Choi EH, Lee HJ. Genetic diversity and molecular epidemiology of the G protein of subgroups A and B of respiratory syncytial viruses isolated over 9 consecutive epidemics in Korea. J Infect Dis 2000; 181: 1547-1556.
- Trento A, Galiano M, Videla C, Carballal G, Garcia-Barreno B, Melero JA, Palomo C. Major changes in the G protein of human respiratory syncytial virus isolates introduced by a duplication of 60 nucleotides. J Gen Virol 2003; 84: 3115-3120.
- Trento A, Viegas M, Galiano M, Videla C, Carballal G, Mistchenko AS, Melero JA. Natural history of human respiratory syncytial virus inferred from phylogenetic analysis of the attachment (G) glycoprotein with a 60-nucleotide duplication. J Virol 2006; 80: 975-984.
- de-Paris F, Beck C, de Souza Nunes L, Machado AB, Paiva RM, da Silva Menezes D, Pires MR, dos Santos RP, de Souza Kuchenbecker R, Barth AL. Evaluation of respiratory syncytial virus group A and B genotypes among nosocomial and community-acquired pediatric infections in Southern Brazil. Virol J 2014; 11: 36.
- Salter A, Laoi BN, Crowley B. Molecular epidemiology of human respiratory syncytial virus subgroups A and B identified in adults with hematological malignancy attending an Irish hospital between 2004 and 2009. J Med Virol 2011; 83: 337-347.
- Yu JR, Kim S, Lee JB, Chang J. Single intranasal immunization with recombinant adenovirus-based vaccine induces protective immunity against respiratory syncytial virus infection. J Virol 2008; 82: 2350-2357.