Research Article - Journal of Plant Biotechnology and Microbiology (2018) Journal of Plant Biotechnology and Microbiology (Special Issue 1-2018)
ePARTS: An easy way to store and access your MoClo construct information in plant biotechnology projects.
Pardo J, Caro E*Centre for Plant Biotechnology and Genomics. Universidad Politécnica de Madrid (UPM) - Instituto Nacional de Investigacióny Tecnología Agrariay Alimentaria (INIA). Campus Montegancedo, 28223, Madrid, Spain
- *Corresponding Author:
- Elena Caro
Center for Biotechnology and Plant Genomics UPM-INIA
Campus Montegancedo, 28223
Madrid
Spain
Tel: +34 913 36 60 00
E-mail: elena.caro@upm.es
Accepted Date: October 11, 2017
Citation: Pardo J, Caro E. ePARTS: An easy way to store and access your MoClo construct information in plant biotechnology projects. J Plant Biotechnol Microbiol. 2017;1(1):1-7.
Abstract
The use of modular cloning techniques like MoClo has become very popular in plant biotechnology projects that deal with the creation of multigenic constructs. The storage of the complex information related to these constructs is absolutely essential and we found that there are no free tools designed specifically for MoClo constructs available. We have designed a relational database that represents the data of interest and how they are related. A program developed in Python programming language - ePARTS - was written to make the database accessible to a broad range of users. It includes a visual interface that allows to insert and search constructs and other items as primers, enzymes or antibiotic resistances in a user friendly way. The source code of the developed system and the database is freely available to allow plant biotech community that deals with complex MoClo constructs benefit from it.
Keywords
Primer, PCR, MoClo(Modular Cloning System), Tkinter (TCL/TK).
Introduction
Synthetic biology is based on the idea that gene networks are formed of modular parts and their different combination allows for the development of artificial regulatory networks with a biotechnological potential. Since its beginnings, scientist tried to introduce two engineering concepts in synthetic biology development: the standardization of parts and the abstraction hierarchy [1,2]. With this idea in mind, “parts” where described with well-defined common features so that their combination could lead to more complex systems [3-7]. BioBricks parts were pioneers in the establishment of a standardized format for genetic parts. They consisted on DNA sequences flanked by restriction sites which could be combined to create more complex assemblies [8]. This method established three levels of hierarchy: part (functional unit), assembly (sum of parts with a specific function) and systems (a group of assemblies). After that, many assembly methods appeared, based on different molecular biology concepts [4], like the methods based on type II restriction enzymes (RE) [9]. Their main disadvantages were the low efficiency (parts are added one by one) and the creation of an 8 bp scar between parts [10].
The methods based on type IIS REs (that cleave outside of their recognition sequence) allow for more freedom in the design of the ends generated after digestion, making possible the assembly of more parts in one single reaction [4]. The parts are obtained through PCR or synthesis [11,12] in such a way that after their digestion the generated ends overlap with those of the adjacent parts. The first method developed using this type IIS REs was Golden Gate [13], but it required that the position of the parts was defined a priori [4]. GoldenBraid [14,15] and MoClo [16,17] are methods for plant synthetic biology based on GoldenGate where this problem has been solved by the creation of position standards. GoldenBraid reduces the number of vectors used, but due to its simplicity it increases the economic and temporal cost in the generation of complex multigenic constructs [14,15,18,19]. MoClo is more versatile and efficient and has become the system of choice for many synthetic biology laboratories.
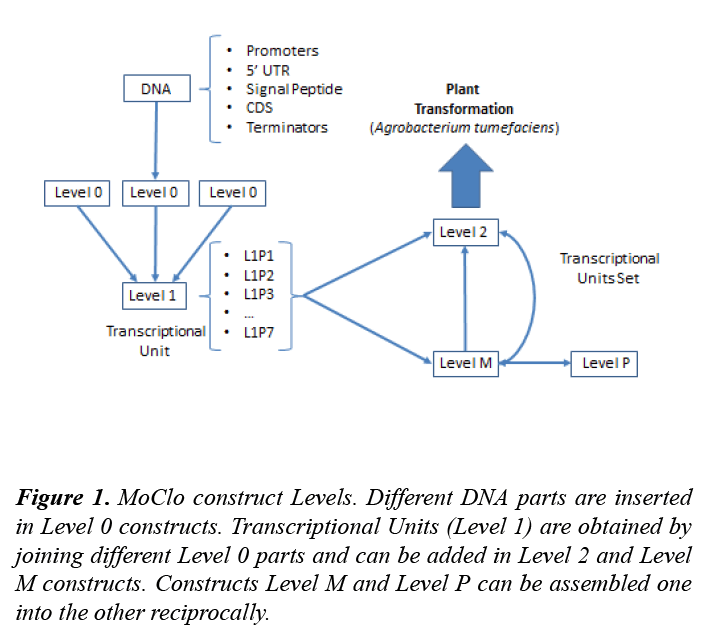
The Modular Cloning System for Standardized Assembly of Multigene Constructs (MoClo) was released in 2011 [16,17]. It establishes five different levels of constructs: Level 0, Level 1, Level 2, Level M and Level P. Level 0 constructs contain elementary BioBrick parts: promoters, coding sequences (CDS), terminators… that can be combined to generate a transcriptional unit (TU) in a Level 1 construct. Each TU can be cloned in seven different positions (L1P1, L1P2… L1P7) that differ in the flanking sequences that will be generated after a digestion. Up to six Level 1 TUs can be assembled in the desired order to create a Level 2 construct. Level 1 constructs can also be inserted in a Level M plasmid and be later added as a whole to a Level 2 construct (Figure 1).
Figure 1: MoClo construct Levels. Different DNA parts are inserted in Level 0 constructs. Transcriptional Units (Level 1) are obtained by joining different Level 0 parts and can be added in Level 2 and Level M constructs. Constructs Level M and Level P can be assembled one into the other reciprocally.
Due to this complexity it becomes necessary to develop tools for the storage of all the information related the constructs generated for synthetic biology projects and that are adapted to the specificities of the cloning system used. We present here a database and associated access program designed ad-hoc for the management of MoClo constructs that will be useful for plant biotechnology laboratories worldwide.
Methods
Database design
The relational database management system (RDBMS) used was MySQL Server v5.7. The RDBMS was deployed in a machine with a static IP used as server in order to allow remote accessing to the database.
Program for database access: The program to access to the database, ePARTS, was written using Python (v. 3.4.0) as programming language. The graphical interface was designed using Tkinter (TCL/TK).
Results
Relational database
The database implemented allowed to manage all the information of the constructs available in the laboratory. Each construct (transformed in E. coli DH5 α) is identified by a single code that matches with the code of the cryogenic tube where the cells are preserved in glycerol (in database terminology, this code was the primary key of the table “glycerol”). This code is an alphanumeric string based on the combination of the first characters (refer to the collection to which the tube belongs) and a number (to identify the sample in the collection). Acceptor vectors are stored and identified the same way since at the collections there is any difference between constructs and the acceptor vectors.
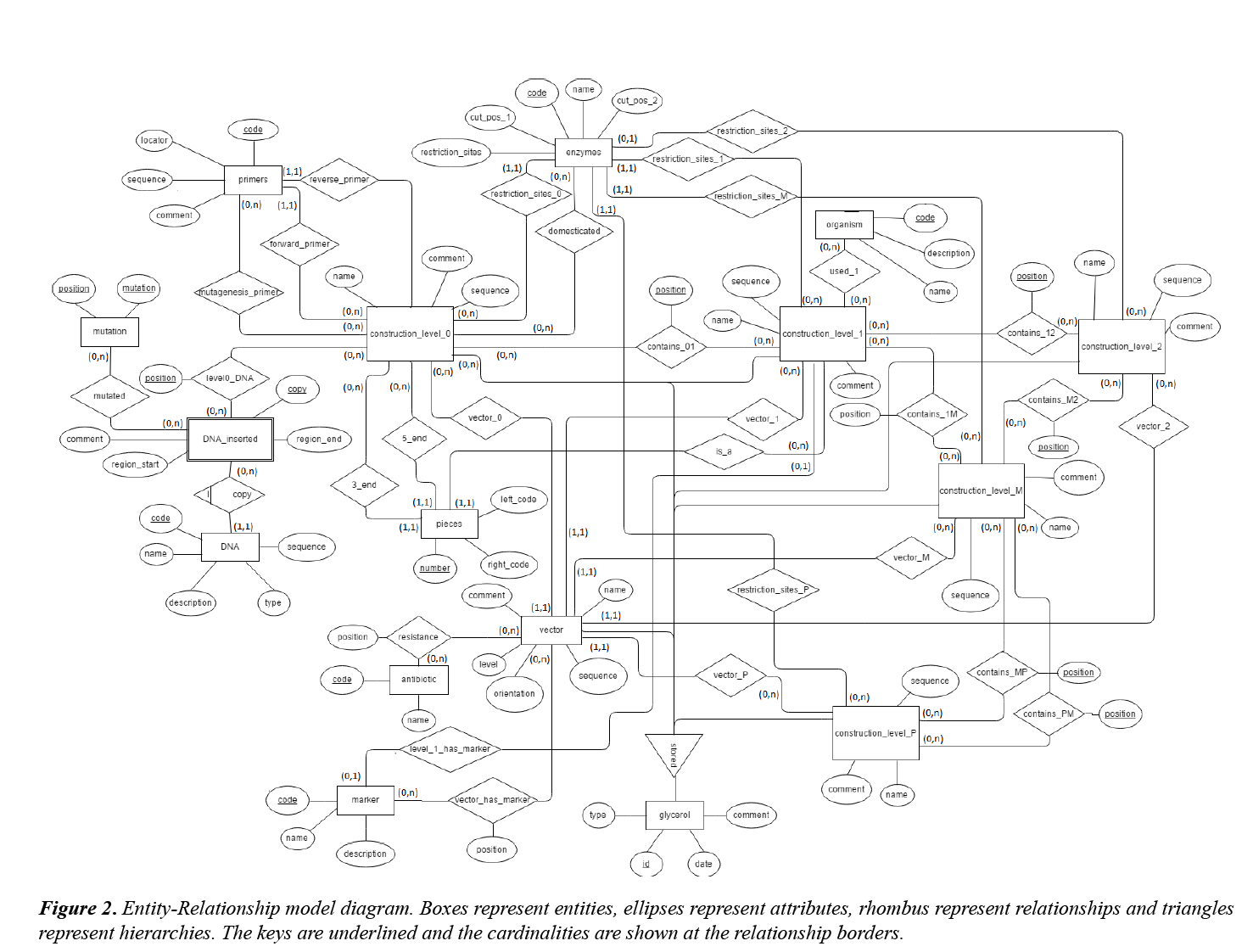
An Entity: Relationship (E-R) model was designed to represent the schema of the data to be stored in the database (Figure 2).
The entity called “glycerol” is the main element of the database and it has as attributes the sample code (“id”), the date when it was added to the collection (“date”) and a comment (“comment”) where information such as the person who deposited it is specified. Entities “construct_level_0”, “construct_level_1”, “construct_level_2”, “construct_level_M”, “construct_level_P” and “vector” refer to the different constructs stored at different MoClo levels and to the available acceptor vectors. All of them are glycerol disjointed subtypes, though they are related with “glycerol” through a hierarchy. Furthermore, these entities have a name ("name"), a nucleotide sequence ("sequence") and a short description ("comment"). The "vector" entity counts also with the attributes "level" (indicates the MoClo level of the constructs that can be cloned inside it) and "orientation" (it only has a value when "vector.level" = "level1" and its possible values are "forward" and "reverse", depending on the orientation of the TU inside the vector). Entities corresponding to constructs are related to "vector" by a (0,n) cardinality for the constructs and a (1,1) cardinality for "vector”, since an acceptor vector can be used to generate different constructs but for each construct, only one vector is used.
"DNA" entity contains the DNA pieces (promoters, CDS, terminators...) considered at the design of the different constructs. This element counts with the attributes "sequence" (nucleotide sequence, usually obtained from GeneBank), "type" (the type of element concerned), “name” (gene or region's name, followed by the organism to which it belongs), “description” and "code" (identifier). However, the elements employed when generating Level 0 constructs are not "DNA" instances but the ones from "DNA_insterted". This is a weak entity dependent of "DNA" whose entries make reference to the concrete sequences that are inserted (they are copies of the "DNA" elements, that can be slightly different from the originals). They are identified by the "DNA.code" and by "copy" (copy number). Its other attributes are "comment" (description), "region_start" and "region_end" (the first and the last nucleotide at the copy, taking as reference the original sequence). "DNA_inserted" is related to the entity "mutation" in order to indicate the sequence variations respect to the "DNA" instance; these changes are identified by the position of the base affected ("position", entity primary key) and the new nucleotide (or nucleotides) placed at that point ("mutation").
"DNA_inserted" is related to "construct_level_0" through "level0_DNA" relationship. This relationship counts with the attribute "position” that determines the order in which the different DNA elements have been inserted in the construct. Due to the fact that the same "DNA_inserted" element (the same "DNA" copy) can be inserted several times in different positions inside the same Level 0 construct, "position" is a primary key. The relationships that relate “construct_level_0” with “construct_level_1”, “construct_level_1” with “construct_level_2” and “construct_level_M”, “construct_level_M” with “construct_level_2” and “construct_level_P” and “construct_level_P” with “construct_level_M” work the same way: all of them are relationships (0,n) for both entities that intervene and in which the "position" attribute is a primary key that indicates the inserts relative order. Do not confuse this position (that only indicates the insertion order) and the different Level 1 positions that are determined by the acceptor vector in which they are cloned.
"Primers" is an entity that includes all the primers employed to generate the different level 0 constructs. They are characterized by a code ("code"), a locator ("locator") that indicates the physical location of the aliquot, their sequence ("sequence") and a description of them ("comment"). "primers" and "construct_level_0" are related by "forward_primer", "reverse_primer" and "mutagenesis_primer" (primer used to introduce punctual mutations) relationships. "forward_primer" and "reverse_primer" have cardinality (1,1) for "primers" (each construct has only one primer of each type) and cardinality (0,n) for "construct_level_0" (the same primer can be employed to generate more than one construct). "mutagenesis_primer" has cardinality (0,n) for both entities.
As already explained in the introduction, MoClo is based on the combination of modules that after being digested with a IIS type RE result in DNA fragments with overlapping ends. The ends that are generated are those that determine their position within the final construct. The "pieces" entity groups the different possible positions identified by a code ("number", Level 1 pieces have codes whose value starts with 'P') and the four bases sequences that characterize them (attributes "right_code" and "Left_code"). The enzymes used in the generation of constructs are included as records of the entity "enzymes". This entity has a code ("code") that identifies the enzyme, a name ("name"), the sequences that it recognizes ("restriction_sites") and the positions in which it cuts the strand with the recognized sequence ("cut_pos_1") and the complementary one ("cut_pos_2"), taking as reference the last base of the recognition sequence. This entity relates to the entities of the different levels of constructs through the relationships "restriction_sites_0", "restriction_sites_1", "restriction_sites_P". These indicate the restriction targets present in the constructs that allow the next level of assembly. These are relationships with cardinality (1,1) for enzymes, less in the case of "restriction_sites_2" where the cardinality is (0,1). This is because Level 2 constructs may not have restriction targets. "Enzymes" is also related to "construct_level_0" through "domesticated"; this relationship indicates enzymes that do not have restriction targets within the insert of the construct. That is, if a construct is domesticated for BsaI (RE), it means that the insert present in that construct does not have targets recognized by that enzyme. This is important because if presented, it could not be used to generate higher levels constructs that require a BsaI digestion.
The "antibiotic" and "marker" entities refer to the different antibiotics and selection markers used in the selection of colonies after the transformations. Both are related to "vector" by relations with cardinality (0, n). These relationships have the attribute "position" indicating whether the antibiotic or marker is inside (value 'IN') or outside (value 'OUT') of the region of the vector that is lost after digestion.
Finally, the entity "organisms" refers to the different organisms in which the constructs can be used (which organisms can express the genes they contain). It relates only to "constructs_level_1", using the "used" relationship, because Level 1 constructs are the lower level constructs with functionality by them.
According to the information provided by the E-R model, the tables that comprise the database were created. 28 tables were generated: glycerol, DNA, DNA_inserted, mutation, level0_DNA, construct_level_0, construct_level_1, construct_level_2, construct_level_M, construct_level_P, vector, contains_01, contains_12, contains_1M , contains_M2 , contains_MP, contains_PM, primers, mutagenesis_primer, pieces, enzymes, domesticated, antibiotics. marker, vector_has_marker, organisms and used. These tables can be found in Annex I. The relationships (1: n) were transformed by the foreign key propagation mechanism. In all the foreign keys it was decided to restrict the deletion of the keys and to change (cascade) their values in case of modifications in the key registries. The primary keys of the tables involved in the hierarchy (the different "glycerol" subtypes) are the "id" from the "glycerol" table (foreign keys). In all tables where a "DNA_inserted" element is referenced, both "DNA" and "copy" codes are part of the primary key; this happens in the tables "mutation" and "level0_DNA".
Tables needed to implement the database in your server are available at GitHub: https://github.com/SilencingLab/e-PARTS/tree/master/database-tables/
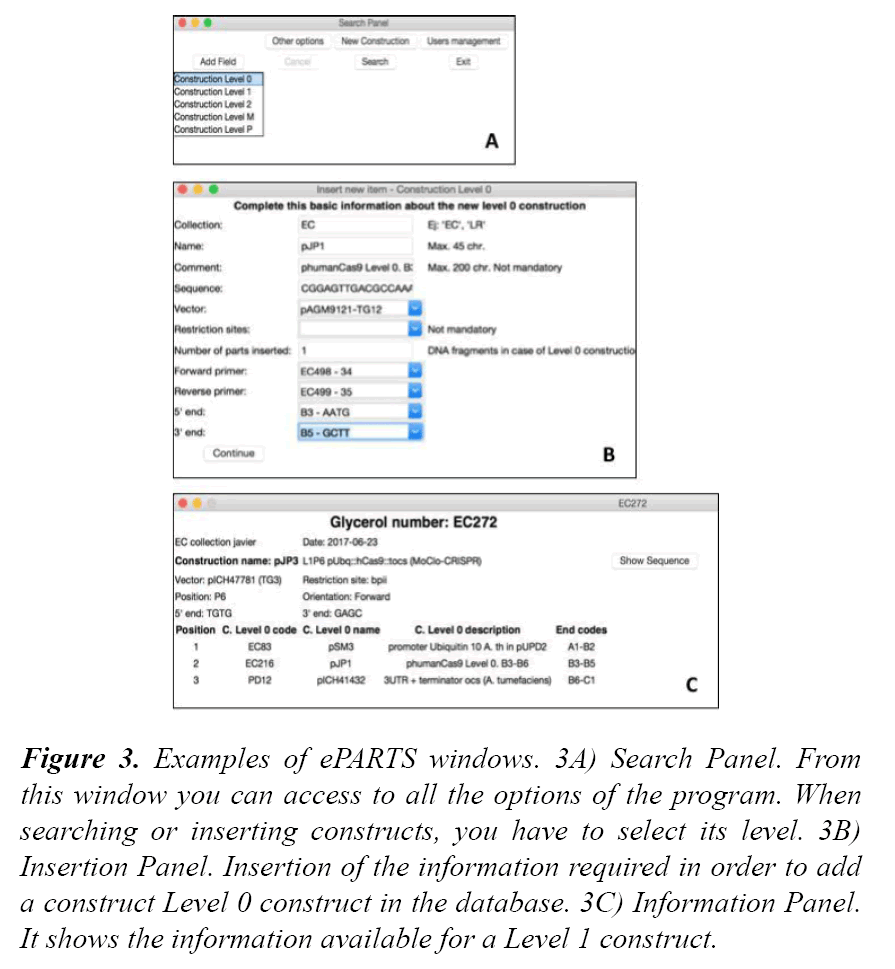
ePARTS was the program developed to interact with the information contained in the database. It provides a graphical user interface that facilitates the search and the insertion tasks of the different constructs. Each window is explained below (Figure 3).
Figure 3: Examples of ePARTS windows. 3A) Search Panel. From this window you can access to all the options of the program. When searching or inserting constructs, you have to select its level. 3B) Insertion Panel. Insertion of the information required in order to add a construct Level 0 construct in the database. 3C) Information Panel. It shows the information available for a Level 1 construct.
Welcome screen: This screen asks for database access credentials (username and password) and the IP address and the schema which is wanted to be accessed. If the credentials are correct, the Welcome Screen is closed and the Search Panel is displayed.
From this window the user can access all the options offered by the program (there is a different button for each one): Search and Insertion of constructs, Search and Insertion of other elements (primers, DNA elements, enzymes ...), User Management and Exit.
Search panel: To search for a construct the user has to indicate the type (level) of the construct he wants to look for. This is done from a list box on the left side of the screen. After selecting it, up to eight filters can be added (by pressing the "Add field" button) to optimize the search. The filters are different depending on the level of construct selected. This is because, for example, a Level 0 construct may be interesting to be looked for by the type of DNA it contains (promoter, CDS ...), whereas in the case of searches of Level 1 constructs it may be interesting to filter the results by its orientation. The different alternatives for each of the levels are in a dictionary whose keys are the possible construct levels. All possible values (present in the database) for those fields are displayed once selected the different filters to be used in the search and after pressing the "Options" button. The results obtained after querying are displayed in different combo boxes, from where the user can choose the desired value. By pressing the "Search" button all the constructs that meet all the selected criteria are obtained (if there were any filter selected, all the constructs of the chosen level would be obtained). The search is done through a single query created by concatenating the different tables and necessary conditions; so the query will be longer the greater the number of filters used. The returned results (constructs that meet the established criteria) are stored in a tuple defined as a global variable. After the search, the Search Panel is closed and the Result Panel is displayed.
To insert a new construct in the database you have to select the level of the construct to be entered in the list box and click on the "New Construct" button, and then the Insertion Panel will be displayed. Unlike what happens during searches, in this case, the Search Panel does not close but is minimized, allowing you to work with both screens at the same time.
The "Other Options" and "Users management" buttons cause the appearance of the Other Options Panel and the Users Panel, respectively, as well as the minimization of the Search Panel. The "Cancel" button is enabled after adding a filter and allows you to reset the Search Panel. The same happens when the close-window button (the "X") is pressed. In order not to restart this screen and to close the application, you have to press the "Exit" button.
Result panel: This screen displays a list of the constructs that have passed the Search Panel filters. The results of this query are stored in the global variable discussed above. It is a tuple of tuples, in which each of the main elements refers to a construct. Each of these main elements is a tuple containing the "code", the "name", the "comment" and the "vector" of the construct. This information is presented in a table and with an option button next to each of the constructs. Each of the buttons has associated a value that corresponds to the "code" of the respective construct. Once an option is selected, the "Show" button is enabled. Pressing this button minimizes the Result Panel and displays the Construct Panel (the construct code is passed as an argument), which contains the selected construct information. Then, the "New Search" allows the Search Panel to be restarted, making it possible to re-query the database and access all the options that the window presents. If the variable "search_result" does not contain any elements, a label that indicates that there are no results for the established search criteria is displayed and the "Show" button is not displayed.
Construct panel: At the start of this screen, the "code" of the selected construct is passed as an argument. Different queries are made with this data with the objective of obtaining all the relevant information about the construct, which is the one shown on the screen. Depending on the type (level) of construct in question, different fields will be displayed. At the top of the screen appears the basic information of the construct: glycerol tube and collection where it is stored, the user who deposited it, construct name and comment. Information such as the vector used, the restriction targets presented, the primers that have been used, their positions... also appears on this panel. Finally, in the lower part of the screen are shown in a table, in order and detailed, the different parts that have been inserted in the construct. For instance, if the information shown corresponds to a Level 2 construct, this section will show the data of the different Levels 1 and Levels M that have been used in its generation. In this screen also the "Sequence" button is displayed (it shows the construct sequence in the command window).
Insertion panel: The content of this window also depends on the level of the construct to be inserted. The page consists of different fields (entries and combo boxes) in which the information of the new construct has to be inserted. The combo boxes values are obtained after several queries, similar to those made in the Search Panel to obtain the possible values of the filters. Pressing the "Continue" button, all the data that has been entered is checked to be correctly inserted and it is added to the database by an "INSERT" query. The glycerol identifier (which identifies the construct) is obtained by consulting the samples already stored in the database belonging to the collection where the new sample will be stored. Once the identifiers of all these samples are obtained, the last one is checked. Adding a unit to the numerical part of it gives the new code. One of the fields to be filled is the number of parts that the construct has (for example, in the case of levels M constructs, how many inserts from Levels 1 or P have been used). The Inserted Constructs Panel will be displayed as many times as this number indicates.
Constructs inserted panel: This screen allows you to select the constructs, or DNA fragments ("DNA_inserted"), in case you are inserting a Level 0 construct, which have been used to generate the new construct. The possible inserts are shown in a combobox whose values are obtained after one or two queries (for example, in the case of introducing a Level P construct, two queries are made since both Level M constructs and Level 1 constructs could have been used to generate it). The first time this window is shown is due to the fact that it has been called from the Insertion Panel, passing as arguments the number of parts to be inserted, the position (within the construct, not the position determined by the ends) of the part corresponding to that iteration, the code of the new construct and its name. The screen will be destroyed and iteratively displayed until the position is equal to the indicated number of parts; in these cases, the call is made from the window itself. In each iteration, the code of the new construct, the code of the inserted construct (newly selected) and the position of this one in the new construct are inserted in the tables that relate the different entities of construct levels ("contains_01" for example). Once all the positions have been covered without any error, the database is committed, and the insertion of the information of the new construct takes place.
Other options panel: From this window, you can manage the information related to the rest of the elements of the database. The screen consists of two comboboxes: the first allows you to select the desired action (search or insertion of elements) and the second, the type of element to be searched or inserted ("DNA", "DNA_inserted", "marker "," Primers "," antibiotic "or" vector ").
If the user chooses the option to search for an element already present in the database, a combobox with all possible values is displayed. For example, if you have selected to search a primer, the combobox will display all the available primers (both codes and locators are shown). Once you have selected the specific item to be searched for and the "Continue" button is pressed, the Other Options screen is closed and the Information Panel is displayed with the data of interest. It happens the same in the case that the desired item is of another type.
Instead, if the user chooses to insert a new element, some entries and combo boxes appear on the screen. There the information necessary to perform the insertions in the database is entered or selected. These fields vary depending on the type of item you have selected. Once they have been completed, the new element is added. In most cases, the generation of a code that identifies the element is required. This is generated from those previously stored in the database, just as it has been discussed in the case of glycerols.
In case the inserted element is "DNA_inserted" or "vector" may appear new windows (Panel Other Options 2). In the case of "DNA_inserted", the new windows will appear if the presence of mutations in the DNA copy has been indicated (there will be as many iterations as indicated mutations). In the case of "vector", these screens allow the selection of resistors and markers. The operation of these windows is similar to that described for the Inserted Building Panel.
Information panel: This screen displays the basic information of the selected item in the Other Options Panel. Depending on the type of element, some fields or others will be displayed. Fields are displayed in labels, while their values are displayed in entries, allowing you to select and copy them to the clipboard of your computer. The information of the different fields is obtained by performing different queries.
Users management panel: From this screen, you can increase the number of users who can access the database. To add a new user, a name and password are requested. You must also choose the level of user privileges. It establishes 4 levels, depending on the actions they have allowed: Level 1 (only search); Level 2 (Level 1+insertions); Level 3 (Level 2+update and delete) and Level 4 (Level 3+user’s management).
The program’s code is available at GitHub: https://github.com/SilencingLab/e-PARTS/tree/master/ePARTS.py
Discussion
Standardization of parts in synthetic biology
One of the current challenges of Synthetic Biology is the establishment of a common format for the different modular parts. The use of this common format has several advantages, such as the possibility of establishing standardized procedures and protocols, allowing to maximize the use of the generated resources, in addition to easing collaboration between groups. Moreover, the standardized format is the only way to efficiently store and manage the information of the constructs that are generated.
An example of this is the program designed and implemented here. This is based on the idea that all generated constructs share a similar format (all constructs are equivalent to an acceptor vector with an insert and have restriction targets that allow the generation of further constructs) and are related to each other (the different DNA elements are inserted in Level 0 constructs, these in Level 1 constructs and so on). These features of the MoClo system make possible the implementation of a database and a program of access to it. Such a task would not have been possible if each of the parts were assembled differently had completely different characteristics to the rest.
Synthetic biology constructs management tools available
There are repositories that allow for the storage of generated constructs, as well as all useful information related to them (acceptor vector, resistance, application), like the Standardized Biological Parts Registry (RSBP) [20], a public repository developed as a digital catalog and physical store of genetic parts in Bio Brick format. Also, some interfaces that allow users to design and assemble genetic circuits already exist. These types of applications simplify the design process by unifying different tools such as design of primers, analysis of sequences and visualization of constructs. Some examples are the Golden Braid website (http://www.gbcloning.org/), Raven (http://www.ravencad.org/) and MoClo Planner [1]. The latter is a specific software designed for the MoClo described in the literature, but for which there is no distribution available to date. ePARTS is the first free code tool designed specifically for plant biotechnology management of MoClo constructs.
User experience
In many labs, the information of the constructs generated is kept in simple ways, such as a spreadsheet. This way, it is not possible to access all the complete information (sequences, primers, resistances), and a lot of it is lost with time. It is very important that all the details of the parts and constructs available are easy to access for everyone since the number of constructs generated is increasing considerably thanks to the simplicity of the experimental procedure necessary to obtain them. With the implementation of ePARTS, it is very easy to describe the basic characteristics of the constructs, since all the relevant information can be consulted at a glance, facilitating both the work of the members of a laboratory and the sharing of resources with the research community.
Acknowledgement
This work was supported by Ramón y Cajal Grant from the Ministry of Economy and Competitiveness of Spain RYC-2012-10367 and State Research Program Scientific and Technical Excellence project BIO2014-58789-P. We thank Prof. Alejandro Rodríguez-González and Prof. Ernestina Menasalvas for solving our doubts during the development of this project and proofreading of the manuscript.
References
- Shaer O, Valdes C, Liu S, et al. MoClo planner: Interactive visualization for Modular Cloning bio-design. BioVis 2013 - IEEE Symposium on Biological Data Visualization. 2013; pp: 57-64.
- Cameron DE, Bashor CJ, Collins JJ. A brief history of synthetic biology. Nat Rev Microbiol. 2014;12:381-90.
- Benner S. Benner, S.A. Synthetic biology: act natural. Nature. 2003;421.
- Casini A, Storch M, Baldwin GS, et al. Bricks and blueprints: methods and standards for DNA assembly. Nat Rev Mol Cell Biol. 2015;16:568-76.
- Monod J, Jacob F. Teleonomic mechanisms in cellular metabolism, growth, and differentiation. Cold Spring Harb Symp Quant Biol. 1961;26:389-408.
- Cookson NA, Mather WH, Danino T, et al. Queueing up for enzymatic processing: correlated signaling through coupled degradation. Mol Syst Biol. 2014;7:561.
- Endy D. Foundations for engineering biology. Nature. 2005;438:449-53.
- Knight T. Idempotent Vector Design for Standard Assembly of Biobricks. MIT Libr. 2003;1-11.
- Cohen SN, Chang ACY, Boyer HW, et al. Construction of Biologically Functional Bacterial Plasmids In Vitro. Proc Natl Acad Sci. 1973;70:3240-44.
- Sleight SC, Bartley BA, Lieviant JA, et al. In-fusion biobrick assembly and re-engineering. Nucleic Acids Res. 2010;38:2624-36.
- Mutalik VK, Guimaraes JC, Cambray G, et al. Precise and reliable gene expression via standard transcription and translation initiation elements. Nat Methods. 2013;10:354-60.
- Linshiz G, Stawski N, Goyal G, et al. PR-PR: Cross-platform laboratory automation system. ACS Synth Biol. 2014;3:515-24.
- Engler C, Kandzia R, Marillonnet S. A one pot, one step, precision cloning method with high throughput capability. PLoS One. 2008;3.
- Sarrion-Perdigones A, Falconi EE, Zandalinas SI, et al. GoldenBraid: An iterative cloning system for standardized assembly of reusable genetic modules. PLoS One. 2011;6.
- Sarrion-Perdigones A, Vazquez-Vilar M, Palaci J, et al. GoldenBraid 2.0: A Comprehensive DNA Assembly Framework for Plant Synthetic Biology. Plant Physiol. 2013;162:1618-31.
- Weber E, Engler C, Gruetzner R, et al. A modular cloning system for standardized assembly of multigene constructs. PLoS One. 2011;6.
- Werner S, Engler C, Weber E, et al. Fast track assembly of multigene constructs using golden gate cloning and the MoClo system. Bioeng Bugs. 2012;3:38-43.
- Binder A, Lambert J, Morbitzer R, et al. A modular plasmid assembly kit for multigene expression, gene silencing and silencing rescue in plants. PLoS One. 2014;9.
- Lampropoulos A, Sutikovic Z, Wenzl C, et al. GreenGate - A novel, versatile, and efficient cloning system for plant transgenesis. PLoS One. 2013;8.
- http://partsregistry.org/Main_Page