Research Article - Journal of Parasitic Diseases: Diagnosis and Therapy (2018) Volume 3, Issue 1
Pfmdr1 gene polymorphism of Plasmodium falciparum isolates from asymptomatic individuals of Dienga, southeastern Gabon
Irène Pegha-Moukandja1,2*, Lady Charlene Kouna2, Sydney Maghendji-Nzdondo1, Jean Claude Biteghe-Bi-Essone1, Dominique Fatima Voumbo-Matoumona1, Fousseyni S Touré Ndouo1, Jean Bernard Lekana-Douki12
1Unité de Parasitologie Médicale (UPARAM), Centre International de Recherches Médicales de Franceville (CIRMF) B.P. 769 Franceville, Gabon
2Département de Parasitologie-Mycologie et Médecine Tropicale, Faculté de Médecine, Université des Sciences de la Santé, B.P. 4009 Libreville, Gabon
- *Corresponding Author:
- Irène Pegha-Moukandja
Unité de Parasitologie Médicale
Centre International de Recherches Médicales de Franceville (CIRMF)
B.P. 769, Franceville, Gabon
Tel: (+241) 06 03 80 06
E-mail: moukandja@hotmail.com
Accepted on March 20, 2018
Citation: Moukandja IP, Kouna LC, Nzdondo SM, et al. Pfmdr1 gene polymorphism of Plasmodium falciparum isolates from asymptomatic individuals of Dienga, southeastern Gabon. J Parasit Dis Diagn Ther. 2018;3(1):18-25. doi: 10.4066/2591-7846.1000024
DOI: 10.4066/2591-7846.1000024
Visit for more related articles at Journal of Parasitic Diseases: Diagnosis and TherapyAbstract
Background: In Gabon, studies on molecular epidemiology of malaria are mainly confined to urban and hospital settings; contrasting with rural areas where data are scarce, especially on asymptomatic P. falciparum infection. In this study, the polymorphism of P. falciparum multidrug resistance -1 gene (Pfmdr1) was investigated for the first time in P. falciparum specimens from asymptomatic individuals living in Dienga, a rural area in southeastern Gabon.
Methods: P. falciparum specimens collected in 2013 and 2014 and diagnosed by standard microscopy (microscopic infection=MI) or by PCR (submicroscopic infection=SMI) were used in this study. The codons 86, 184 and 1246 of Pfmdr1 of P. falciparum specimens were genotyped by PCR and RFLP.
Results: Among the three codons 86, 184 and 1246 of Pfmdr1, the wild-type alleles were the most prevalent in 2014. The frequency of mixed genotype N86Y was 19.23% in 2013 and 25.28% in 2014. The frequency of D1246Y mixed genotype was 10.40% in 2013 and 17.24% in 2014. On the other hand, no mixed genotype was observed for the codon 184. The most frequent haplotypes identified were NFY (33%) in 2013 and YYD (30.0%) in 2014. The frequencies of other haplotypes (YYY, YFD, NYD, YFY, NFD and NYY) did not show significant difference between 2013 and 2014.
Conclusion: Data from Dienga showed moderate variations in the distribution of Pfmdr1 genotypes and a high haplotype diversity suggesting the low ACT drug pressure on parasites in this locality.
Keywords
Asymptomatic P. falciparum infection, Rural Gabon, Pfmdr1 polymorphism.
Background
Malaria still caused 429 000 (235 000–639 000) deaths around the world; from which 78% occurred in sub-Saharan Africa [1]. This infection can unpredictably progress to severe forms such as severe anemia and cerebral malaria which are responsible for almost all the morbidity and mortality related to the disease [2- 5]. The emergence of antimalarial drug-resistance is one of the causes of this disease’s persistence.
In 2000, the Roll Back malaria initiative has permitted to the international community to intensify the measures to fight malaria. As a result, a marked decline of mortality and morbidity due to malaria was observed. This achievement is owed to a rise in investments for vector control such as the use of Insecticide Treated Bed Nets (ITNs), and treatment including Intermittent Preventive Treatment during pregnancy (IPTp) and combined therapies, especially Artemisinin-Based Combined Therapies (ACTs) against uncomplicated malaria [6-8]. However, these successes are threatened by the emergence of parasite resistance against artemisinin [9-11]. Moreover, this resistance was firmly established in Cambodia, Thailand, Vietnam and Burma and its possible spread over the rest of South-east Asia has been documented [12,13]. On the other hand, the therapeutic failures of ACTs could be associated with single nucleotide polymorphism (SNP) in P. falciparum multidrug resistance gene-1 (Pfmdr1) [14,13], as well as with K13 propeller [15,16]. Pfmdr1 is an ATP cassette protein located on the parasite’s food vacuole. Indeed, the mutations in this gene reduce the parasite’s sensitivity to anti-malarial drugs [17]. The therapeutic failures following ACTs treatments were associated with the selection of N86, 184F and D1246 (NFD genotype), in the Pfmdr1 gene [18]. The NFD haplotype was also involved in the decrease of Artemether-Lumefantrine (AL) sensibility. Also, N86, Y184 and D1246 (NYD), might be implicated in the failure of the Amodiaquine treatment (Amodiaquine is used in combination with Artesunate). Furthermore, an increase of asexual parasites and gametocytes harboring the Pfmdr1 NFD haplotype in patients treated with AL was linked with treatment failure [19].
In Gabon, located in West-Central Africa, an equatorial climate prevails. Seasonality is characterized by two rainy seasons and two dry seasons. Malaria transmission is perennial and the resistance to CQ, amodiaquine and sulfadoxine-pyrimethamine is widespread [20-23]. In 2003, in line with WHO recommendations, the National Malaria Control Program adopted the ACT strategy to treat uncomplicated malarial spraying, instead of CQ and other monotherapies. The ACTs most widely used in Gabon are artemether-lumefantrine (AL) and artesunate-amodiaquine (AS-AQ). Consequently, the prevalence of malaria among febrile children fell from 30% to 13% between 2004 and 2008, the mean age of children with malaria rose from 24 to 41 months [21,7]. Later, in 2013, several studies demonstrated the rise of malaria in urban areas of the country (Libreville, Oyem and Franceville) [24,25]. In addition, an increased prevalence of the Pfmdr1 86N genotype of P. falciparum from symptomatic individuals was observed in Franceville [26], confirming previous data from Oyem (North of Gabon) [27]. All these previous studies were conducted in urban hospitals and were mainly performed on symptomatic individuals By contrast; little is known about malaria epidemiology in rural areas.
Dienga is a village located in the south-east of Gabon near the Congo border which counts almost 3,000 inhabitants.
The aim of this study was to determine the prevalence of polymorphisms in Pfmdr1 gene associated with parasites resistance to antimalarials of P. falciparum isolated from asymptomatic individuals of Dienga.
Methods
Study site
Dienga is a densely forested rural area, situated near the Congo border, with around 3,000 inhabitants. As in most parts of the country, malaria is highly endemic and P. falciparum is predominantly due to Anopheles gambiae (80%), with an entomological infection rate of 100 infective bites per human per year [28]. Malaria seasonal peaks of transmission coincide with the rains, from February to June and from September to December [28].
Ethical approval
This study was approved by the National Ethics Committee of Gabon and registered under number (N°0018/2013/SG/CNE). Written informed consent was obtained from participants, parents or legal guardians upon enrollment in the study.
Study population and biological samples
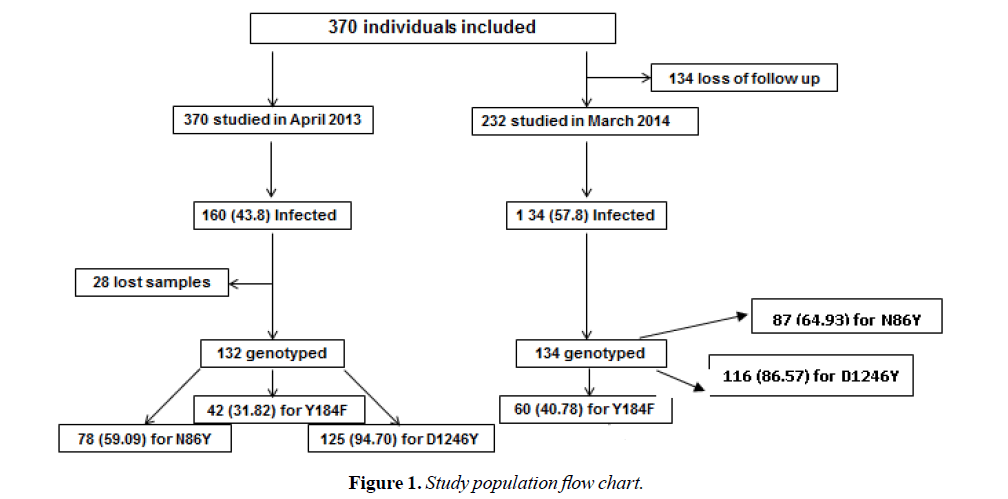
From April 2013 to June 2014, through six field missions, a cohort composed of 370 indiduals was followed to investigate P. falciparum infection [29]. The participants were composed of volunteers permanently living in Dienga; primary school children (≥ 3 years) and adults. Following physical examination, the auxiliary temperature and history of fever (≥ 37.5°C) in the previous 24 hours for all participants were recorded. Blood samples were collected by venipuncture in 5-ml EDTA tubes during the first mission (April 2013), the fifth (March 2014) and the last (June 2014). P. falciparum infection was diagnosed during each mission using microscopy and PCR (STEVOR gene) to detect microscopic (MI) and submicroscopic infections (SMI) respectively.
Overall, 266 P. falciparum positive blood samples (132 isolates: 76 MI and 56 SMI, in April 2013 and 134 isolates: 59 MI and 75 SMI in March 2014) were taken to determine Pfmdr1 polymorphism. Samples were then composed of a total of 135 MI and 131 SMI (Table 1 and Figure 1).
| Baseline characteristic | Apr-13 | Mar-14 |
|---|---|---|
| n | 370 | 232 |
| Means Age (yrs.) ± SD | 24.34 (22.65) | 21.87 (20.77) |
| Males (%) | 184 | 115 |
| Females (%) | 186 | 117 |
| Distribution by age (%) | ||
| 3-6 yrs | 31 | 23 |
| 7-10 yrs | 100 | 68 |
| 11-14 yrs | 67 | 48 |
| 15-18 yrs | 36 | 20 |
| 19 + yrs | 136 | 73 |
Table 1. Baseline Characteristics of the study subjects.
P. falciparum DNA extraction and analysis of Pfmdr1 polymorphism
DNA extraction was performed from the 266 P. falciparum isolates (microscopic and submicroscopic) using the DNeasy Blood & Tissue kit according to the manufacturer's procedure (QIAGEN, Hilden, Germany). Pfmdr1 polymorphism was assessed using PCR-RFLP as previously described [26]. Primer sequences for N86Y, D184F and D1246Y SNPs PCR and restriction enzymes are shown in Table 2 [19].
| Genes, Codons | Primer names | Nucleotide sequence | T°C | Restriction enzyme | Sizes (bp)* |
|---|---|---|---|---|---|
| Pfmdr1, N86Y | mdr86D1 | TTTACCGTTTAAATGTTTACCTGC | 45 | Afl III | 126 +165 |
| mdr86D2 | CCATCTTGATAAAAAACACTTCTT | ||||
| Pfmdr1, D1246Y | mdr1246D1 | AATGTAAATGAATTTTCAAACC | 45 | Bgl II | 126 +165 |
| mdr1246D1 | CATCTTCTCTTCCAAATTTGATA | ||||
| Pfmdr1, Y184F | mdr184A | AAAGATGGTAACCTCAGTATCAAAGAAGAG | 40 | Dra I | 114 +356+528 |
| mdr184B | AAAGATGGTAACCTCAGTATCAAAGAAGAG | 114+242+173+32 |
Table 2. Sequences of primer sets, and restriction enzymes used to characterize polymorphisms.
Data processing and analysis
Statistical analysis was performed using STATA 14.0 (Texas 77845 USA, 4905 Lakeway Drive College Station). The Chi-square test was used to compare categorical variables among groups. The non-parametric K-wallis, Pearson’s and Fisher’s exact tests were used for group comparisons, as appropriate. First, a descriptive analysis was done for the study population. Then, univariate analysis was achieved to investigate the link between Pfmdr1 alleles and both microscopic and submicroscopic infections. In all statistical tests, significance was assumed at p<0.05.
Results
Distribution of PfMDR1 polymorphism over the study period
This study investigated the occurrence of Pfmdr1 alleles (codons 86, 184 and 1246) in P. falciparum isolates from asymptomatic individuals of Dienga, southeastern Gabon. Overall, 266 isolates were included, 132 in 2013 and 134 in 2014.
Out of the 132 P. falciparum isolates from 2013, 78 (59.00%), 42 (31.82%) and 125 (94.70%) were successfully genotyped for N86Y, Y184F and D1246Y SNPs respectively in 2013. Among the 134 P. falciparum isolates from 2014, 87 (64.93%) for N86Y, 60 (44.78%) for Y184F and 116 (86.57%) for D1246Y were genotyped.
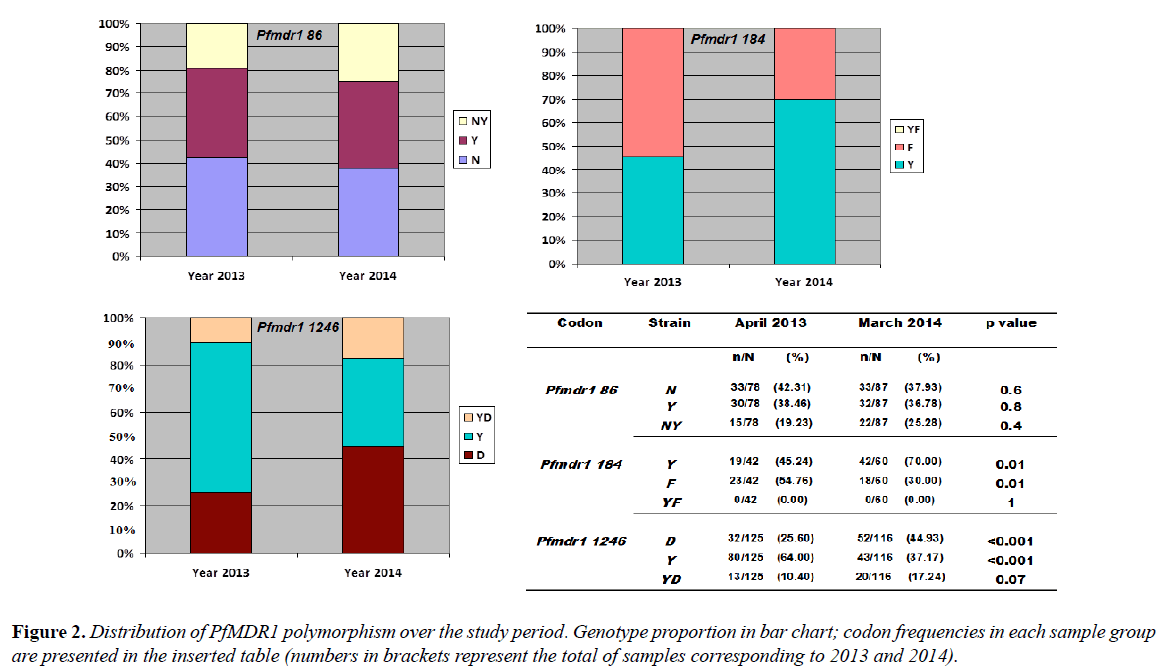
Among the 78 P. falciparum isolates of 2013, Pfmdr1 codon 86 typing gave 33 (42.31%) wild-type genotypes, 30 (38.46%) mutated types and 15 (19.23%) mixed genotypes. For Pfmdr1 codon 184, 19/42 (45.24%) wild genotypes and 23/42 (54.76%) of the 184F mutated genotype were observed. The codon 184 did not show mixed genotypes. The Pfmdr1 codon 1246 genotype determination gave 32/125 (25.60%) of the D1246 wild type, 84/125 (64.0%) of the 1246Y mutated type and 13/125 (10.40%) of mixed genotypes (Figure 2).
Of the 87 P. falciparum isolates analyzed in 2014, the Pfmdr1 codon 86 showed 33 (37.93%) of the N86 wild type, 32 (36.78%) of the 86Y mutated type and 22 (25.28%) of mixed genotypes. For codon 184, 42/60 (70.0%) were of the Y184 wild type and 18/60 (30.0%) were of the 184F mutated type. No mixed genotype was obtained. Pfmdr1 codon 1246 was composed of 52/116 (44.93%) of D1246, 43/116 (37.17%) of 1246Y and 20/116 (17.24%) of mixed genotype D1246Y (Figure 2).
When P. falciparum isolates from 2013 were compared to those from 2014 there was no significant temporal variation in the individual distribution of SNPs at codon 86 (for N86 p=0.6; 86Y p=0.8 and for N86Y p= 0.4). Nevertheless, the mutated types 1246Y (64% in 2013 vs. 37.17% in 2014) and 184F (54.76% vs 30.0%) were significantly higher in 2013 than in 2014 with p<0.001 and p=0.01 respectively. There was no significant difference in the proportions of the D1246Y mixed genotype between the two sampling times and no Y184F mixed genotype was observed in codon 184 during the study (Figure 2).
Haplotypes
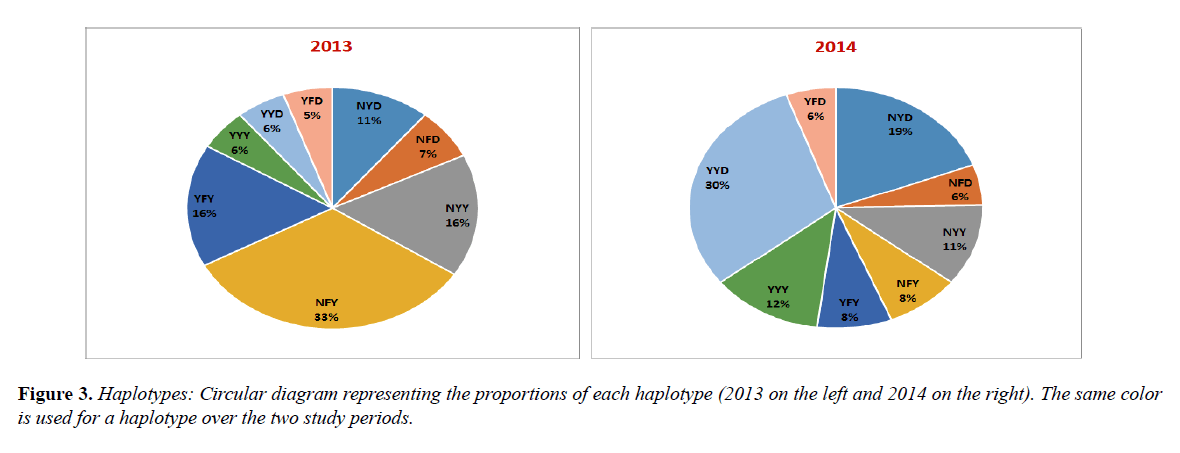
Pfmdr1 three codon-basis haplotypes were compared according to the sampling periods. While the frequency of NFY decreased considerably from 33.0% in 2013 to 8.0% in 2014 (p<0.001), YYD increased significantly from 6.0% in 2013 to 30.0% in 2014 (p=0.01).
The frequencies of the others haplotypes, YYY (6.0% vs. 12.0%), YFD (5.0% vs 6.0%), NYD (11.0% vs 19.0%), YFY (16.0% vs. 8.0%) and NFD (7.0% vs 6.0%) and NYY (16.0% vs 11.0%), were not statistically different over the study periods (Figure 3).
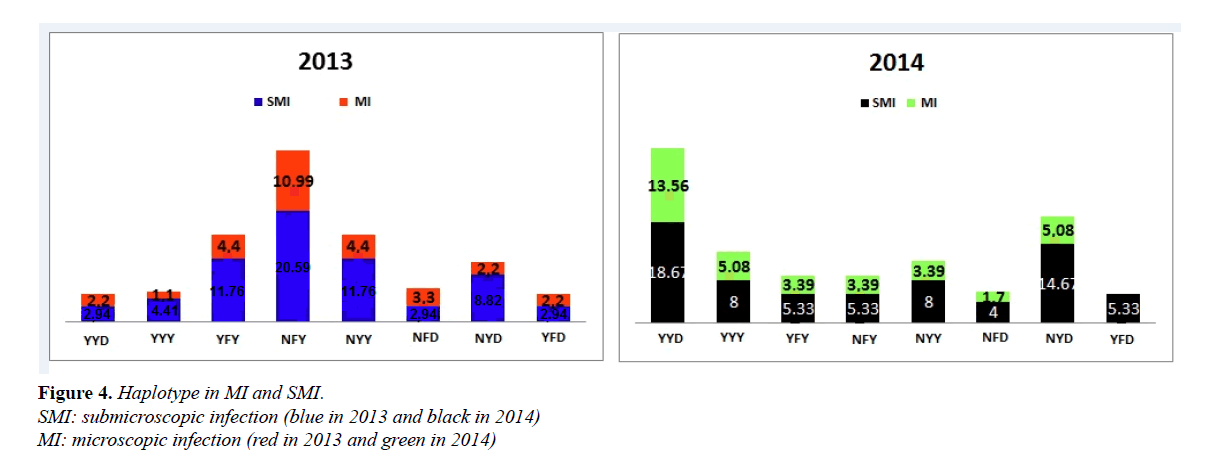
In 2013, the proportions of NYY in SMI were 11.76% against 4.44% for MI isolates, YFY (11.76% SMI vs 4.44% MI) and NFY (20.59% SMI vs 10.99% MI). These results show that these proportions were significantly higher for SMI compared to MI (respectively p=0.04, p=0.04 and p=0.03). Similarly, in 2014, the NYD haplotype was higher in SMI than in MI (8.0% vs 3.39%) (p=0.03); (Figure 4).
Comparing 2013 and 2014 haplotypes according to the infection type, YYD and NYD from SMI significantly increased in 2014 compared to 2013 (p=0.003 and p=0.04), (Figure 4).
Haplotypes in MI and SMI
Some haplotypes tended to be associated with either MI or SMI. These associations were more perceived in the YYY and NYD haplotypes. SMI parasites tended to show almost fourfold (4) the risk of carrying haplotypes YYY and NYD when compared to non-SMI parasites; contrasting with MI isolates in which this risk is very low (Table 3).
| Haplotype | SMI+ (n) | OR (95% CI) | P value | MI+ (n) | OR (95% CI)2 | P value |
|---|---|---|---|---|---|---|
| YYD | ||||||
| Positive | 14 | 1.46 (0.58-3.68 | 0.43 | 8 | 0.75 (0.27-1.72) | 0.43 |
| Negative | 8 | 1 | 14 | 1 | ||
| YYY | ||||||
| Positive | 6 | 1.62 (0.42-6.19) | 0.5 | 3 | 0.62 (0.16-2.37) | 0.5 |
| Negative | 3 | 1 | 6 | 1 | ||
| YFY | ||||||
| Positive | 4 | 1.60 (0.33-2.16) | 0.59 | 2 | 0.62 (0-3.03) | 0.59 |
| Negative | 2 | 1 | 4 | 1 | ||
| NFY | ||||||
| Positive | 4 | 1.60 (0.33-2.16) | 0.59 | 2 | 0.62 (0-3.03) | 0.59 |
| Negative | 2 | 1 | 4 | 1 | ||
| NYY | ||||||
| Positive | 6 | 2.48 (0.55-2.11) | 0.26 | 2 | 0.40 (0-1.83) | 0.26 |
| Negative | 2 | 1 | 6 | 1 | ||
| NFD | ||||||
| Positive | 3 | 2.41 (0.33-2.43) | 0.44 | 1 | 0.41 (0-2.99) | 0.44 |
| Negative | 1 | 1 | 3 | 1 | ||
| NYD | ||||||
| Positive | 1 | 3.21 (0.91-11.19) | 0.07 | 3 | 0.31 (0.09-1.1) | 0.07 |
| Negative | 3 | 1 | 1 | 1 | ||
| YFD | ||||||
| Positive | 11 | 0.24 (0.02-0.53) | 0.07 | 3 | 0 (0-19) | 0.07 |
| Negative | 3 | 1 | 11 | 1 | ||
Table 3. Analyses of associations between haplotypes and either submicroscopic or microscopic infections in 2014
NFD was the sole haplotype which tended to be associated with MI parasites (Table 3). In 2014, the analysis of this association gave the same results.
Discussion
In Dienga, the prevalence of P. falciparum infection decreased around 2003 [30] but a decade after this prevalence rose markedly [29]. This situation may be associated with a lack of ongoing efforts to fight against the disease (ie local anti-malarial drug use, prevention measures). A surveillance of molecular markers of drug resistance, in parasites from asymptomatic individuals, a hidden reservoir, is particularly important to improve malaria control strategies. In sub-Saharan African countries, the monitoring of malaria and anti-malarial drug resistance in rural areas is unusual, as most data are commonly reported from urban areas in Gabon [24,26], in Zanzibar [31], Cameroon [32], Senegal [33] and Mozambique [34].
Two decades ago, chloroquine (CQ) and sulfadoxine pyrimethamine (SP) were the most widespread anti-malarial drugs used in Gabon. Because of the lack of the efficacy of CQ and SP to treat malaria they were replaced by ACT in 2005 by the Gabonese Malaria National Control Program (MNCP) which launched a nationwide campaign of ITNs distribution, intermittent preventive treatment (IPT), and educational messages. The use of artesunate-amodiaquine (AS-AQ), and artemether-lumefantrine (AL) were therefore recommended in treatment of uncomplicated malaria.
The aim of the current study was to investigate the baseline of polymorphisms in the resistance-associated genes Pfmdr1 of parasites collected from asymptomatic individuals of Dienga, considering both microscopic and submicroscopic infections. To our knowledge it is the first study to analyze Pfmdr1 or other gene polymorphism of P. falciparum from non-febrile individuals in a rural area of Gabon.
In this study, codon 1246 of Pfmdr1 was the most efficiently genotyped (94.7%) in 2013 and (86.57%) in 2014 compared to other codons (or alleles). This could be due to either the low parasite density as most of the isolates were submicroscopic and from asymptomatic individuals, or to the variability of P. falciparum field isolates. This result could also be due to the sensitivity of the 1246 primers which may be more sensitive than those of codons 86 and 184 even when the amounts of DNA are low. This result is similar to that observed in Franceville by a team that investigated the Pfmdr1 gene polymorphism [35]. Like Dienga, the city of Franceville is also located in the south-east of Gabon and their similar epidemiological contexts could explain this similarity.
The wild types N86 and D1246 were found at 42.31% and 25.60% respectively in 2013, at 37.93% and 44.93% in 2014. This result shows a relative stability of N86 over the study period. This may be due to the level of ACTs pressure on local parasites which presumably is low in rural areas such as Dienga. However one year is too short a time to appreciate drug pressure impact.
The increase of genotype N86 has been associated with a decreased susceptibility to artemisinins in vitro [36,37]. Our results did not confirm the study of Lekana-Douki et al, which reports a rise in the prevalence of genotype N86 in Franceville after the use of AS-AQ [24] and a study conducted in Mozambique [38].
A relative high prevalence of mutant types 184F and 1246Y in samples from 2013 (54.76 and 64.0%, respectively) was observed. These prevalences significantly dropped in 2014 (30.0 and 37.17% respectively). For codon 1246, this is consistent with data from Mozambique, where the prevalence of genotype 1246Y declined from 5.8% to 2.4%, from 2003- 2005 to 2010-2012 after the treatment with AL [38]. For allele 184, data showed a decrease of mutated genotype 184F between 2013 to 2014 contrasting with Tanzanian data which showed the increase of 184F from 10% to 30% in 2006 to 2010 after AL use [39,36].
A moderate level of the mutated genotype 86Y observed in this study may be associated with CQ resistance; even Pfcrt locus has been not investigated. Moreover, in a study performed in Uganda, it has been documented that genotype 86Y was more important in asymptomatic carriers compared to febrile ones [36]. It has been previously documented that Pfmdr1 86Y and 1246Y mutations decrease chloroquine and amodiaquine sensitivity, but they increase lumefantrine, mefloquine and dihydroartemisin sensitivity according to Pickard [13,40,36,41].
All eight Pfmdr1 haplotypes (codons 86, 184 and 1246) were found at diverse levels, in P. falciparum isolates from Dienga, both in 2013 and 2014. This finding suggests that the high diversity in rural areas may be due to the low drug pressure, contrasting with urban areas of Sudan where only two haplotypes have been found [42]. Interestingly, the prevalence of the NFD haplotype, strongly involved in the decrease of AL sensitivity, was one of the lower haplotypes in the study and that could indicate a small consumption of AL in Dienga. Nonetheless, this result contrasted with data from six cities in Tanzania, where NFD was one of the most common haplotypes encountered after AL treatment [43].
The YYD haplotype significantly increased in 2014, in contrast to Kenya and Tanzania where a decrease of Lumefantrine susceptibility was reported followed by increase of the prevalence of NFD, NYD, YYY and YYD [44,36]. A rise of YYD in 2014 was observed and is consistent with the work of Morris U et al. who reported that YYD is the most prevalent haplotype in asymptomatic malaria infection [45].
Conclusion
In Dienga, P. falciparum from asymptomatic individuals showed moderate variations in the distribution of Pfmdr1 alleles despite the fact that it carried all the Pfmdr1 haplotypes. Likewise, the low prevalence of most of these haplotypes may suggest a limited ACT drug pressure.
Acknowledgment
We thank Dr R.F Kassa Kassa and J Ondzanga for their help as physicians. We are grateful to the individuals and their families who accepted to participate in this study, and to the staff of the CIRMF in Dienga, especially Helene Tiga and Suzan Moundemba. We also express our gratitude to Justice Mayombo, Lewobo Ludjer, Omnella Mavoungou, Ingrid Ontsia and Dr Mpiga-Mickoto. Finally, we thank the staff of the Centre International de Recherches Médicales de Franceville (CIRMF), for their help in the lab tests, particularly Jeanne Lahonko and Dr Statiana-Mbouyi.
References
- Agnandji ST, Fernandes JF, Bache EB, et al. Clinical development of RTS, S/AS malaria vaccine: a systematic review of clinical Phase I-III trials. Future Microbiol. 2015;10(10):1553-78.
- Bouyou-Akotet MK, Nzenze-Afene S, Ngoungou EB, et al. Burden of malaria during pregnancy at the time of IPTp/SP implementation in Gabon. Am J Trop Med Hyg. 2010;82(2):202-9.
- Bouyou-Akotet MK, Mawili Mboumba DP, Kendjo E, et al. Anaemia and severe malarial anaemia burden in febrile Gabonese children: a nine-year health facility based survey. J Infect Dev Ctries. 2013;7(12):983-9.
- Bouyou-Akotet MK, Offouga CL, Mawili-Mboumba DP, et al. Falciparum malaria as an emerging cause of fever in adults living in Gabon, Central Africa. Biomed Res Int. 2014;2014:351281.
- Marsh K, Otoo L, Greenwood BM. Absence of crisis form factor in subjects immune to Plasmodium falciparum in The Gambia, West Africa. Trans R Soc Trop Med Hyg. 1987;81(3):514-5.
- Sy O, Cisse B, Tairou F, et al. Acceptability of indoor residual spraying in the Central-Western of Senegal. Bull Soc Pathol Exot. 2015;108(3):213-7.
- Bouyou-Akotet MK, Mawili-Mboumba DP, Kendjo E, et al. Evidence of decline of malaria in the general hospital of Libreville, Gabon from 2000 to 2008. Malar J. 2009;8:300.
- Ishengoma DS, Mmbando BP, Segeja MD, et al. Declining burden of malaria over two decades in a rural community of Muheza district, north-eastern Tanzania. Malar J. 2013;12:338.
- Dondorp AM, Nosten F, Yi P, et al. Artemisinin resistance in Plasmodium falciparum malaria. N Engl J Med.2009; 361(5):455-67.
- Lim P, Alker AP, Khim N, et al. Pfmdr1 copy number and arteminisin derivatives combination therapy failure in falciparum malaria in Cambodia. Malar J. 2009;8:11.
- Mounanga M, Revangue BS. HIV-1 seroprevalence in pregnant women in Libreville (Gabon). J GynecolObstet Biol Reprod. 1993;22(5):509-12.
- Wongsrichanalai C, Pickard AL, Wernsdorfer WH, et al. Epidemiology of drug-resistant malaria. Lancet Infect Dis.2002;2(4):209-18.
- Pickard AL, Wongsrichanalai C, Purfield A, et al. Resistance to antimalarials in Southeast Asia and genetic polymorphisms in pfmdr1. Antimicrob Agents Chemother. 2003;47(8):2418-23.
- Mita T, Tanabe K. Evolution of Plasmodium falciparum drug resistance: implications for the development and containment of artemisinin resistance. Jpn J Infect Dis. 2012;65(6):465-75.
- Isozumi R, Uemura H, Kimata I, et al. Novel mutations in K13 propeller gene of artemisinin-resistant Plasmodium falciparum. Emerg InfectDis. 2015;21(3):490-2.
- Straimer J, Gnadig NF, Witkowski B, et al. Drug resistance. K13-propeller mutations confer artemisinin resistance in Plasmodium falciparum clinical isolates. Science. 2015;347(6220):428-31.
- Cowman AF, Karcz S, Galatis D, et al. A P-glycoprotein homologue of Plasmodium falciparum is localized on the digestive vacuole. J Cell Biol. 1991;113(5):1033-42.
- Baliraine FN, Rosenthal PJ. Prolonged selection of pfmdr1 polymorphisms after treatment of falciparum malaria with artemether-lumefantrine in Uganda. J Infect Dis. 2011;204(7):1120-4.
- Happi CST, Gbotosho GO, Folarin OA, et al. Selection of Plasmodium falciparum multidrug resistance gene 1 alleles in asexual stages and gametocytes by artemether-lumefantrine in Nigerian children with uncomplicated falciparum malaria. Antimicrob Agents Chemother. 2009;53(3):888-95.
- Aubouy A, Bakary M, Keundjian A, et al. Combination of drug level measurement and parasite genotyping data for improved assessment of amodiaquine and sulfadoxine-pyrimethamine efficacies in treating Plasmodium falciparum malaria in Gabonese children. Antimicrob Agents Chemother. 2003;47(1):231-7.
- Nsimba B, Guiyedi V, Mabika-Mamfoumbi M, et al. Sulphadoxine/pyrimethamine versus amodiaquine for treating uncomplicated childhood malaria in Gabon: a randomized trial to guide national policy. Malar J. 2008;7:31.
- Ndong JM, Atteke C, Aubouy A, et al. In vitro activity of chloroquine, quinine, mefloquine and halofantrine against Gabonese isolates of Plasmodium falciparum. Trop Med Int Health. 2003;8(1):25-9.
- Ramharter M, Wernsdorfer WH, Kremsner PG. In vitro activity of quinolines against Plasmodium falciparum in Gabon. Acta Trop. 2004;90(1):55-60.
- Biteghe Bi Essone JC, Lékana-Douki JBI, Toure-Ndouo FS, et al. Submicroscopic Infection from uncomplicated Plasmodium falciparum malaria of Franceville, southeasern Gabon. Int J Adv Res. 2014;2(1):117-23.
- Mawili-Mboumba DP, Bouyou Akotet MK, Kendjo E, et al. Increase in malaria prevalence and age of at risk population in different areas of Gabon. Malar J. 2013;12:3.
- Lekana-Douki JB, Dinzouna Boutamba SD, Zatra R, et al. Increased prevalence of the Plasmodium falciparum Pfmdr1 86N genotype among field isolates from Franceville, Gabon after replacement of chloroquine by artemether-lumefantrine and artesunate-mefloquine. Infect Genet Evol. 2011;11(2):512-7.
- Mawili-Mboumba DP, Ndong Ngomo JM, Maboko F, et al. Pfcrt 76T and pfmdr1 86Y allele frequency in Plasmodium falciparum isolates and use of self-medication in a rural area of Gabon. Trans R Soc Trop Med Hyg. 2014;108(11):729-34.
- Elissa N, Migot-Nabias F, Luty A, et al. Relationship between entomological inoculation rate, Plasmodium falciparum prevalence rate, and incidence of malaria attack in rural Gabon. Acta Trop. 2003;85(3):355-61.
- Pegha Moukandja I, Biteghe Bi Essone JC, Sagara I, et al. Marked Rise in the Prevalence of Asymptomatic Plasmodium falciparum Infection in Rural Gabon. PLoS One. 2016;11(5):e0153899.
- Toure FS, Mezui-Me-Ndong J, Ouwe-Missi-Oukem-Boyer O, et al. Submicroscopic Plasmodium falciparum infections before and after sulfadoxine-pyrimethamine and artesunate association treatment in Dienga, Southeastern Gabon. Clin Med Res. 2006;4(3):175-9.
- Sisowath C, Ferreira PE, Bustamante LY, et al. The role of pfmdr1 in Plasmodium falciparum tolerance to artemether-lumefantrine in Africa. Trop Med Int Health. 2007;12(6):736-42.
- Eboumbou Moukoko EC, Bogreau H, Briolant S, et al. Molecular markers of Plasmodium falciparum drug resistance. Med Trop (Mars). 2009;69(6):606-12.
- Fall B, Pascual A, Sarr FD, et al. Plasmodium falciparum susceptibility to anti-malarial drugs in Dakar, Senegal, in 2010: an ex vivo and drug resistance molecular markers study. Malar J. 2013;12:107.
- Raman J, Mauff K, Muianga P, et al. Five years of antimalarial resistance marker surveillance in Gaza Province, Mozambique, following artemisinin-based combination therapy roll out. PLoS One. 2011;6(10):e25992.
- WHO. World Malaria Report. 2017.
- Mwai L, Kiara SM, Abdirahman A, et al. In vitro activities of piperaquine, lumefantrine, and dihydroartemisinin in Kenyan Plasmodium falciparum isolates and polymorphisms in pfcrt and pfmdr1. Antimicrob Agents Chemother. 2009;53(12):5069-73.
- Bivigou-Mboumba B, Francois-Souquiere S, Deleplancque L, et al. Broad Range of Hepatitis B Virus (HBV) Patterns, Dual Circulation of Quasi-Subgenotype A3 and HBV/E and Heterogeneous HBV Mutations in HIV-Positive Patients in Gabon. PLoS One. 2016;11(1):e0143869.
- Lobo E, de Sousa B, Rosa S, et al. Prevalence of pfmdr1 alleles associated with artemether-lumefantrine tolerance/resistance in Maputo before and after the implementation of artemisinin-based combination therapy. Malar J.2014;13:300.
- Thomsen TT, Ishengoma DS, Mmbando BP, et al. Prevalence of single nucleotide polymorphisms in the Plasmodium falciparum multidrug resistance gene (Pfmdr-1) in Korogwe District in Tanzania before and after introduction of artemisinin-based combination therapy. Am J Trop Med Hyg. 2011;85(6):979-83.
- Rosenthal PJ. The interplay between drug resistance and fitness in malaria parasites. Mol Microbiol. 2013;89(6):1025-38.
- Mosha JF, Sturrock HJ, Greenhouse B, et al. Epidemiology of subpatent Plasmodium falciparum infection: implications for detection of hotspots with imperfect diagnostics. Malar J. 2013;12:221.
- Gadalla NB, Adam I, Elzaki SE, et al. Increased pfmdr1 copy number and sequence polymorphisms in Plasmodium falciparum isolates from Sudanese malaria patients treated with artemether-lumefantrine. Antimicrob Agents Chemother. 2011;55(11):5408-11.
- Straimer Kavishe RA, Paulo P, Kaaya RD, et al. Surveillance of artemether-lumefantrine associated Plasmodium falciparum multidrug resistance protein-1 gene polymorphisms in Tanzania. Malar J. 2014;13:264.
- Malmberg M, Ferreira PE, Tarning J, et al. Plasmodium falciparum Drug Resistance Phenotype as Assessed by Patient Antimalarial Drug Levels and Its Association With pfmdr1 Polymorphisms. J Infect Dis. 2013;207(5):842-7.
- Morris U, Xu W, Msellem MI, et al. Characterising temporal trends in asymptomatic Plasmodium infections and transporter polymorphisms during transition from high to low transmission in Zanzibar, 2005-2013. Infect Genet Evol. 2015;33:110-7.