Research Article - Biomedical Research (2018) Volume 29, Issue 19
Down syndrome: a cytogenetic study in North Indian population
Priyanka Pandey1*, Rakesh Kumar Verma1, Navneet Kumar1 and Sciddhartha Koonwar2
1Department of Anatomy, King George’s Medical University, Lucknow, India
2Pediatrics, King George’s Medical University, Lucknow, India
- *Corresponding Author:
- Priyanka Pandey
Department of Anatomy
King George’s Medical University, India
Accepted on March 16, 2018
DOI: 10.4066/biomedicalresearch.29-18-415
Visit for more related articles at Biomedical ResearchAbstract
Down syndrome constitutes the most common chromosomal abnormality among live births (1 in 730 live births) and most frequent form of intellectual disability. Genetic cause for this syndrome is trisomy of chromosome 21. The cytogenetic profile of Down syndrome includes free trisomy 21, Robertsonian translocations, mosaicism, duplication of the DS critical region and other structural rearrangements involving chromosome 21. Purpose of the study was to confirm the clinical diagnosis of suspected cases of Down syndrome by karyotyping and to evaluate several risk factors associated with trisomy 21 among patients belonging to Lucknow region. Karyotype analysis was done in 46 patients with clinical suspicion of Down syndrome. GTG banding was done according to standard protocols. Findings: out of 46 cases i.e., 43 (93.5%) had abnormal karyogram. All these 43 cases had trisomy 21 (Down's syndrome). Among patients with Down syndrome, free trisomy (n=40; 93%) was most common followed by Robertsonian translocation (n=2; 4.7%) and mosaic trisomy (n=1; 2.3%) respectively. Majority of patients were males (n=27; 62.7%) and there were 16 (37.3%) females. Male to female ratio of Down syndrome patients was 1.68. 50% of patients with Down syndrome were born to mothers falling in age group of 36-40 y. Identification of various types of chromosomal abnormalities in Down syndrome is very important. It aids in management of these children and to aware the affected families about recurrence risk and options available.
Keywords
Down syndrome, Trisomy 21, Cytogenetic, Karyotyping.
Introduction
Down Syndrome (DS) is the most common chromosomal disorder in humans. It was first described by John Langdon Down in 1866 [1]. Since it is caused by an extra copy of chromosome 21, it is also called trisomy 21 syndrome [2]. Trisomy 21 constitutes the most frequent form of intellectual disability caused by a chromosomal aberration [3-5]. The cytogenetic profile of down syndrome includes free trisomy 21, Robertsonian translocations, mosaicism, duplication of the DS critical region and other structural rearrangements involving chromosome 21 [6-8]. Most common karyotype encountered in Down syndrome children is of free trisomy 21, while their parents have normal karyotype. This type of trisomy 21 is a result of nondisjunction of homologous chromosomes 21during gametogenesis or during early embryonic development after fertilization [9]. Analysis of chromosome heteromorphisms and other informative markers of DNA polymorphisms of patients with Down syndrome and their parents revealed that chromosome 21 nondisjunction occurs more often during the gametogenesis in females than in males [10,11]. Investigations revealed that an extra chromosome 21 mainly originates from errors in maternal meiosis in approximately 90% of the Down syndrome cases and associated with maternal age>35 y [12,13]. In 5-10% of Down syndrome cases the extra chromosome 21 originates due to errors in paternal meiosis, while in less than 5% of cases it results from nondisjunction of chromosomes during a postzygotic mitosis in early embryonic development [14,15]. 4% of DS cases involve Robertsonian translocation in which chromosome 21 may be translocated to another acrocentric chromosome. DS due to translocation can be de novo or inherited from a balanced carrier parent [16]. In general, Robertsonian translocations carriers are phenotypically normal. In 50% of these cases of the rearrangements occur de novo [17] and 95% of the de novo cases originate at the time of maternal meiosis. Around 3-5% of DS cases occur due to mosaicism which involves a non-disjunction postzygotic event. Peoples with mosaic Down syndrome have two distinct cell lines with different karyotype. Some cells have a total of 46 chromosomes with normal karyotype, while others have trisomy in chromosome 21 [18]. There are various conserved features occurring in all DS population, including learning disabilities, craniofacial abnormality and hypotonia in early infancy. Some people of DS are affected by variant phenotypes including Atrioventricular Septal Defects (AVSD) in heart, leukemias (both Acute Megakaryoblastic Leukemia (AMKL) and Acute Lymphoblastic Leukemia (ALL)), AD and HD. DS individual have variety of physical characteristics like a small chin, slanted eye, poor muscle tone, a flat nasal bridge, a single crease of the palm and a small mouth and large protruding tongue. Other features include big toe, abnormal pattern of fingerprint and short fingers [19].
Confirmation of trisomy syndromes is done by cytogenetic techniques. Cytogenetics is the study of the structure and properties of chromosomes, chromosomal behavior during somatic cell division in growth and development (mitosis) and germ cell division in reproduction (meiosis), chromosomal influence on the phenotype and the factors that cause chromosomal changes [20]. Cytogenetic techniques include karyotyping, fluorescent in situ hybridization techniques and Microarray. For routine analysis, however, the classical karyotyping technique using trypsin and Giemsa became the most accepted worldwide. The banding pattern enabled the detection of various structural aberrations like translocations, inversions, deletions, and duplications in addition to the wellknown numerical aberrations [21]. Pinkel et al, 1986 developed a method to visualise chromosomes using fluorescent-labelled probes called Fluorescent in situ Hybridisation (FISH) [22]. FISH technology permits the detection of specific nucleic acid sequences in morphologically preserved chromosomes, cells, and tissues. Using FISH, cytogeneticists can detect chromosomal abnormalities that involve small segments of DNA if their probe is situated, fortuitously or by design, in the affected chromosomal segment [23].
The purpose of this study was to carry out a comparative cytogenetic evaluation of suspected cases of Down syndrome in both sexes using classical karyotyping and to report the incidence of Down syndrome and the frequency of the 3 cytogenetic variants of Down syndrome in Lucknow region, as well as to evaluate the effect of maternal age on the prevalence of Down syndrome.
Material and Method
46 suspected cases of Down syndrome were referred to our laboratory for cytogenetic analysis. These patients showed the clinical features consistent with trisomy 21. Cytogenetic analysis was done by classical karyotyping technique.
For classical karyotyping technique Peripheral venous blood of the patient was collected in BD Vacutainer sodium heparin vial. 0.5 ml of blood sample was taken in 5 ml of culture media (PB MAX) in a test tube, under laminar air flow to maintain sterile condition and this culture tube was incubated at 37°C temperature, 85% humidity and 5% concentration of CO2 for 72 h in slanting position in CO2 incubator (YORCO). After the incubation 5 drops of karyoMAX colcemid solution (0.1 mg/ml) was added, and test tube was centrifuged at 1000 rpm for 10 min. The supernatant was discarded by pipetting of media, leaving behind as little medium as possible over the bottom of test tube. The cell button was suspended in 5 ml of hypotonic solution (potassium chloride+sodium citrate) and was incubated at 37°C for 30 min. The centrifugation was done at 1000 rpm for 10 minutes. Leaving the cell button was resuspended in 5 ml of fixative (methanol+acetic acid). This process of centrifugation, discarding the supernatant and adding the fixative was repeated 2-3 times until button at the bottom of the test tube became white. Finally, the tube was kept at 2-3°C for 2 h before the harvested blood cells became ready for slide preparation. Slides were prepared by dropping method, and were treated with trypsin to obtain better banding. Adequately aged slides were stained with Giemsa stain. These slides were observed under microscope (Olympus BX51) attached with a computer and fields showing a good spared of metaphase were photographed.
Karyograms were prepared from slides using cytovision software. Analysis for various chromosomal aberrations was done with Karyogram. Karyotyping results were obtained by analyzing 20 metaphase fields for each case and in cases where abnormal karyotypes were suspected, the observation was extended to a total of 30 fields. The karyotypes were reported as per international system for human cytogenetic nomenclature (ISCN, 2013) guidelines.
Results
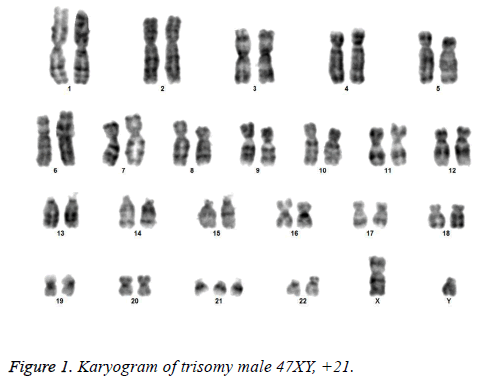
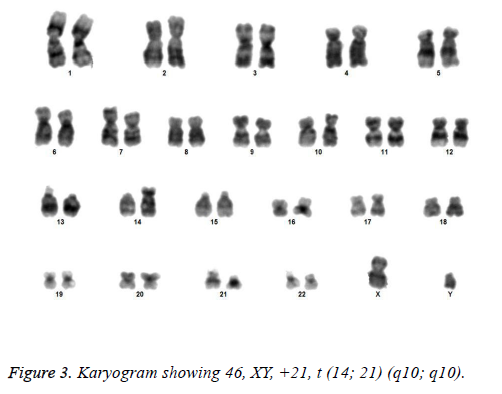
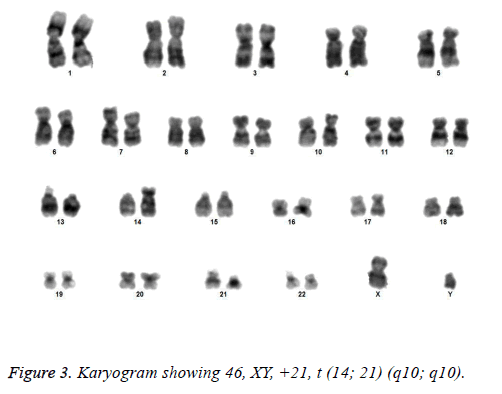
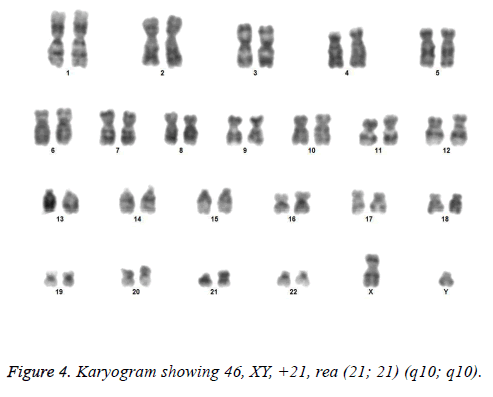
All the patients suspected to have trisomy 21 had physical features of Down syndrome like epicanthic eye fold, brush field spot, flat nasal bridge, abnormal teeth, furrowed tongue, narrow palate, short neck, short and broad hands, incurved 5th finger, gap between 1st and 2nd toes, murmur, muscular hypotonia, oblique eye fissure, blepharitis, conjunctivitis, nystagmus, mouth permanently open, protruding tongue, higharched palate, folded ear, loose neck of skin, short 5th finger and transverse palmar crease. All of the children had mental retardation and 25 cases i.e. 54.3% had congenital heart disease. Average age of children suspected of having DS was 5.6 y. Out of 46 suspected cases, 43 cases showed cytogenetic findings consistent with Down syndrome. On classical karyotyping free trisomy (n=40; 93%) was most common followed by Robertsonian translocation (n=2; 4.7%) and Mosaic trisomy (n=1; 2.3%) respectively (Table 1). Among 40 cases with free trisomy, 24 (55.8%) were males (47XY, +21) and 16 (37.2%) were females (47XX, +21) (Figures 1 and 2). Out of 2 cases with Robertsonian translocation 1 (2.3%) case each was of genotype 46, XY, +21, REA (21; 21) (q10; q10) and 46XY, +21, t (14,21) (q10; 10) (Table 2). Both the patients were male (Figures 3 and 4). The single case with mosaicism was also male.
| Gender | Free trisomy | Robertsonian translocation | Mosaic |
|---|---|---|---|
| Male | 24 (55.8%) | 2 (4.65%) | 1 (2.32%) |
| Female | 16 (37.2%) | 0 | 0 |
Table 1. Distribution of chromosomal abnormalities according to gender (N=43).
| SN | Genotype | No. |
|---|---|---|
| 1 | Free trisomy | |
| 47XY, +21 | 24 | |
| 47XY, +21 | 16 | |
| 2 | Robertsonian translocation | |
| a) | 46, XY, +21, rea (21; 21) (q10; q10) | 1 |
| b) | 46, XY, +21, t (14; 21) (q10; q10) | 1 |
| 3 | Mosaic 47XY, +21/46XY | 1 |
Table 2. Genotype of Down’s syndrome (N=43).
As far as the maternal age was concerned among 40 mothers of trisomy 21 only patients, maximum (n=22; 55%) were aged 36-40 y followed by 31-35 y (30%), <30 y (10%) and >40 y (5%) respectively (Table 3). Among those showing trisomy with Robertsonian translocation-there was 1 (33.3%) mother each in age group <30 years and 31-35 y respectively. In single case of mosaic, the maternal age was in age group 36-40 y.
| Age group (Y) | Free trisomy | Robertsonian translocation | Mosaic |
|---|---|---|---|
| ≤ 30 Y | 4 | 1 | 0 |
| 31-35 Y | 12 | 1 | 0 |
| 36-40 Y | 22 | 0 | 1 |
| >40 Y | 2 | 0 | 0 |
Table 3. Distribution of chromosomal abnormalities according to maternal age at birth (N=43).
Discussion
Cytogenetic analysis done in different population worldwide indicate that free trisomy 21 is the most common variant of DS whose incidence varies between 95.51 to 83.82% as reported by various authors [1]. Our reported incidence i.e. 93% also falls in the same range. The difference in percentages may arise due to the number of metaphases analysed. In our study Robertsonian translocations made the second most frequent variant (4.7%) whose incidence falls within the previously reported range (2.66-5.1%). In Robertsonian translocation most frequently the exchange occurs in non-homologous forms involving two different acrocentric chromosomes-either two different D group chromosomes (chromosomes 13-15), two different G group chromosomes (21 and 22), or a D group and a G group chromosome. 50% of these translocations are de novo and 50% are inherited from a balanced carrier parent (usually the mother). The most common translocation involved in DS is between 14 and 21 which was followed by translocation between two chromosome 21 [24]. In our study we identified 1 case each of rob (14; 21) and rea (21; 21). Our results are also in congruence with reports that identified above mentioned rearrangements as the most frequent translocations associated with trisomy 21. Both the translocations were denovo as the parents of these patients had a normal karyotype. Mosaic DS followed RT in terms of incidence i.e. 2.3% which also is within the range reported from different parts of the world (1.19-10.78%) [1]. This discrepancy can also be explained by number of metaphases analysed in each study.
Advanced maternal age is one of the most important risk factors contributing to the non-disjunction of chromosome. The current study also demonstrated a very strong association of advanced maternal age with the birth of a DS child. The mean maternal age in our study was 35 ± 4.4 y. 50% DS children were born to mothers above 35 y of age, and among the 40 mothers of trisomy 21 only patients, maximum (55%) were aged 36-40 y. This association between increased maternal age and the risk of having a child with Down syndrome was is approved by various studies [25]. Belmokhtar et al. also show very similar results where 54.5% of all DS belonged to free trisomy DS were born to mothers who were in the advanced age group (≥ 35 y) [26]. Flores et al. also reported that there is an increase in birth of free trisomy DS children to mothers above 35 years of age [2]. A study on the prevalence of DS in Atlanta conducted by Siffel et al. during 1994-1999 revealed that the prevalence of DS was 55.3 per 10,000 for women above 35 years age compared to 8.5 per 10,000 for women below 35 years [27]. A study conducted on 52,965 amniocentesis to determine the maternal age of some major chromosomal aberrations suggested that the rate of trisomy 21 increases from maternal age of 35 years [28]. A study in England and Wales showed that mother’s delayed childbearing has led to an increase in the prevalence of DS [29]. Even though, the association between increasing maternal age and the conception of trisomies has been recognized since many years, but the underlying mechanism behind the maternal age effect still remains poorly understood. Increasing rate of meiotic errors due to the aging process of the ovary (biological aging) is one of the many hypotheses given [30]. The long arrest of the oocytes in prophase I of meiosis seems to be underlying mechanism. In females, the entry of the oocytes into meiosis starts during fetal life which is then arrested in a prophase stage of meiosis in which they persist from late fetal life until the time of ovulation. The first meiotic division MI is completed in the female just prior to ovulation and the second division MII is completed only if the egg is fertilized. In female the process takes years to complete [30]. Hence studies from various population groups show that risk of nondisjunction increases with advancing maternal age (>35 y).
Two children belonging to Robertsonian translocation group were born to mothers belonging to age group<30 y and 31-35 y. Hulten et al. hypothesized that most females might be having low grade trisomic 21 ovarian mosaics with an average of 0.54% trisomy 21 cells and concluded that this ovarian mosaicism may predispose to trisomy 21 conception and conception of a translocation DS foetus in a mother at a younger age as compared to free trisomic DS [31]. As in children having RT the peripheral blood lymphocytes from both the parents had normal karyotype so we can conclude that the trisomy 21 due to Robertsonian translocation must have arisen either de novo or due to ovarian mosaicism. Hence, this explains the comparatively young maternal age for translocation trisomic children.
Conclusion
In this study we tried to carry out a comparative cytogenetic evaluation of suspected cases of Down syndrome in both sexes using classical karyotyping and Fluorescent in situ Hybridization (FISH) techniques, to report the incidence of Down syndrome and the frequency of the 3 cytogenetic variants of Down syndrome in Lucknow region, as well as to evaluate the effect of maternal age on the prevalence of Down syndrome. Our study concluded that free trisomy made the most common cytogenetic variant followed by Robertsonian translocation and mosaic trisomy. This study also showed that 50% of children with DS were born to mothers above 35 years of age. Cytogenetic analysis is the most important diagnostic tool for DS. Cytogenetic data also provide a basis for the genetic counselling of families into which DS children are born and further management of children with trisomy 21.
Acknowledgement
Authors are grateful to the head, faculty members and staff of department of anatomy King George’s Medical University for providing support and infrastructure to carry out this work.
References
- Flores-Ramírez F, Guerrero CP, Delgado CG, Morales-Jimenez AB, Arias-Villegas CM, Cervantes A. Cytogenetic profile in 1,921 cases of trisomy 21 syndrome. Arc Med Res 2015; 46: 484-489.
- Lejeune J, Turpin R, Gautier M. Le mongolisme premier exempled aberration autosomique humaine. Ann Genet 1959; 1; 41-49.
- Reeves RH, Baxter LL, Richtsmeier JT. Too much of a good thing: mechanisms of gene action in Down syndrome. Trends Genet 2001; 17: 83-88.
- Chandra N, Cyril C, Lakshminarayana P. Cytogenetic evaluation of down syndrome: A review of 1020 referral cases. Int J Hum Genet 2010; 10: 87-93.
- Sheets KB, Crissman BG, Feist CD. Practice guidelines for communicating a prenatal or postnatal diagnosis of Down syndrome: recommendations of the national society of genetic counselors. J Genet Couns 2011; 20: 432-444.
- Sherman SL, Freeman SB, Allen EG. Risk factors for nondisjunction of trisomy 21. Cytogenet.Genome Res 2005; 111: 273-280.
- Olson LE, Roper RJ, Sengstaken CL. Trisomy for the Down syndrome critical region necessary but not sufficient for brain phenotypes of trisomic mice. Hum Mol Genet 2007; 16: 774-782.
- Gardner RJM, Sutherland R, Shaffer LG. Chromosome abnormalities and genetic counseling (4th Edn.) New York Oxford University Press 2011; 353.
- Petersen MB, Mikkelsen M. Nondisjunction in trisomy 21: origin and mechanisms. Cytogenet Cell Genet 2000; 91: 199-203.
- Antonarakis SE. Parental origin of the extra chromosome in trisomy 21 as indicated by analysis of DNA polymorphisms. N Engl J Med 1991; 324: 872-876.
- Petersen MB, Schinzel AA, Binkert F, Tranebjaerg L, Mikkelsen M, Collins FA. Use of short sequence repeat DNA polymorphisms after PCR amplification to detect the parental origin of the additional chromosome 21 in Down syndrome. Am J Hum Genet 1991; 48: 65-71.
- Mikkelsen M, Hallberg A, Hanne P. Epidemiology study of Downs syndrome in Denmark, including family studies of chromosomes and DNA markers. Dev Brain Dysfunct 1995; 8: 4-12.
- Morris JK, Alderman, Mutton D. Cytogenetic and epidemiological findings in Down syndrome: England and Wales 1989-2009. Am J Med Genet A 2012; 158: 1151-1157.
- Hassold T, Sherman S. Down syndrome: genetic recombination and the origin of the extra hromosome 21. Clin Genet 2000; 57: 95-100.
- Hassold T, Hunt P. To err (meiotically) is human: the genesis of human aneuploydy. Nat Rev Genet 2001; 2: 280-291.
- Grouchy J, Turleau C. Autosomal disorders. Principles and practice of medical genetics. New York Churchill Livingstone 1990: 24771.
- Schaffer LG, Jackson-Cook CK, Stasiowski BA, Spence JE, Brown JA. Parental origin determination in thirty de novo Robertsonian translocations. Am J Med Genet 1992; 43: 957-963.
- Devlin L, Morrison PJ. Mosaic Downs syndrome prevalence in a complete population study. Arch Dis Child 2004; 89: 1177-1178.
- Sinet PM, Theopile D, Rahmani Z, Chettouch Z, Blovin JL, Prier M. Mapping of Down syndrome phenotype on chromosome 21 at the molecular level. Biomed Pharmacother 1994; 48: 247-252.
- Hare WCD, Singh EL. Cytogenetics in animal reproduction. Slough Commonwealth Agricultural Bureaux UK 1979.
- Smeets DF. Historical prospective of human cytogenetics: from microscope to microarray. Clin Biochem 2004; 37: 439-446.
- Pinkel Gray JW, Trask B, Van- den Engh G, Fuscoe J, Van D. Cytogenetic analysis by in situ hybridization with fluorescently labeled nucleicacid probes. Cold Spring Harbor Symp Quant Biol 1986; 51: 151157.
- Dewald GW, Brockman SR, Paternoster SF, Bone ND, OFallon JR, Allmer C. Chromosome anomalies detected by interphase fluorescence in situ hybridization: correlation with significant biological features of B-cell chronic lymphocytic leukaemia. Br J Haematol 2003; 121: 287-295.
- Vikraman SK, Chandra V, Balakrishanan B, Batra M, Kuriakose R, Kannoly G. A rare balanced parental t (21q; 21q) Robertsonian translocation that results in Down syndrome in all viable pregnancies. Int J Reprod Contracept Obstetr Gynecol 2017; 4: 514-517.
- Allen EG, Freeman SB, Druschel C, Hoobs CA, OLeary LA, Romitti PA. Maternal age and risk for trisomy 21 assessed by the origin of chromosome nondisjunction: a report from the Atlanta and National Down Syndrome Projects. Human Gene 2009; 125: 41-52.
- Belmokhtar F, Belmokhtar R, Kerfouf A. Cytogenetic study of down syndrome in algeria: report and review. J Genet Syndr Gene Ther 2016; 7: 280.
- Siffel C, Correa A, Cragan J, Alverson CJ. Prenatal diagnosis, pregnancy terminations and prevalence of Down syndrome in Atlanta. Birth Defects Res A Clin Mol Teratol 2004; 70: 565-571.
- Ferguson-Smith MA, Yates JR. Maternal age specific rates for chromosome aberrations and factors influencing them: report of a collaborative European study on 52 965 amniocenteses. Prenat Diagn 1984; 4: 5-44.
- De Souza E, Alberman E, Morris JK. Down syndrome and paternal age, a new analysis of case-control data collected in the 1960s. Am J Med Genet 2009; 149: 1205-1208.
- Hunt PA. Meiosis in mammals: recombination, non-disjunction and the environment. Biochem Soc Transact 2006; 34: 574-577.
- Hulten MA, Jonasson J, Nordgren A, Iwarsson E. Germinal and somatic trisomy 21 mosaicism: how common is it, what are the implications for individual carriers and how does it come about? Curr Genom 2010; 11: 409-419.