Research Article - Biomedical Research (2017) Volume 28, Issue 3
Docking analysis of some flavanols as Cdk-4 inhibitor as a possible option to treat retinoblastoma
Ye-ping Han1, Ru-wei Chen2 and Feng Zhao3*1Department of Neurosurgery, Henan Province Hospital of Traditional Chinese Medicine, China
2Department of Neurosurgery, Binzhou City of Shandong Province People's Hospital Neurosurgery, China
3Department of Ophthalmology, Dezhou Municipal Hospital, China
Accepted on September 13, 2016
Abstract
In present scenario, the developing economies are struggling with the burden of various forms of cancers. Among them, the heritable form of Retinoblastoma (RB) also known as common intraocular malignancy is considered as a most severe form of cancer affecting mainly children’s. Due to involvement of Cyclin Dependent Kinases (Cdks) in the phosphorylation step, its inhibition offers selective advantage in the prevention of tumour progression by activating the suppressor activity of RB protein. Encouraged by the above, the present study was intended to elucidate the effect of some which are well known for its anticancer potential on the CDK-4 with the help of molecular docking analysis.
Keywords
Retinoblastoma, Docking, Flavanol.
Introduction
In present scenario, the developing economies are struggling with the burden of various forms of cancers [1-3]. Among them, the heritable form of Retinoblastoma (RB) also known as common intraocular malignancy is considered as a most severe form of cancer affecting mainly children’s [4]. It is believed as a rare form for malignant disease affecting only single individual in 15,000 live births. The disease is further classified as unilateral, which identified nearly at two-year after the birth and bilateral has been diagnosed before attaining the age of one year. Despite of its hereditary nature, till now no single evidence has been reported that, it has been predisposed to any specific race or gender. In its early days of discovery in late eighteenth century, the individuals diagnosed with this dreadful disease are not survived much [5-9]. But thanks to the, advanced technology and treatment modalities, which make this dream achievable and prolong the survivability of the affected person.
Chemotherapy is most widely used modalities apart from cryotherapy, laser photocoagulation and plaque brachytherapy for the treatment of RB [10-13]. It has been well known that, Retinoblastoma protein (pRB), a tumour suppressor protein is highly over expressed in many of malignancies including RB. It causes to check excessive cell growth through interfering with the progression of cell cycle till the cell is viable to divide into other cells. During the process of cell division, the pRb is inactivated by phosphorylation and allow the division to progress [14-16]. Due to involvement of Cyclin Dependent Kinases (Cdks) in the phosphorylation step, its inhibition offers selective advantage in the prevention of tumour progression by activating the suppressor activity of pRB. As per the previous study, it has been found the CDK4 has been play a key role in the phosphorylation of pRB and emerged as a prospective target to treat RB [17,18].
Prompted by the above, the present study was intended to elucidate the effect of some flavan-3-ols which are well known for its anticancer potential on the CDK-4 with the help of molecular docking analysis [19].
Results and Discussion
Docking is the most powerful and widely accepted tool to predict the effect of designed ligand with the protein of interest. It provides the appropriate and considerable data for the molecule docked at the active, before it is actually synthesized and tested. This tool is virtually creating the environment where the small molecule is allowed to interact with the protein at the molecular level. It allows the individual to interpret the behaviour of the small molecule in the binding site of the target proteins and helps in understanding the pathway by which the molecule acts. Owing to this capability, various studies are utilizing the approach of drug discovery to prevent iteration in the later stages of discovery. The study has been chiefly rely on two fundamental steps, the first step corresponds to the prediction of conformation of ligand including its position within the binding pocket, whereas, the last step comprise of determination of binding affinity. The later step has been calculated based on the empirical scoring function with the help of certain mathematical algorithms, which provide the exact nature of binding in a quantitative manner. Generally the determination of the binding affinity has been identified on the basis of six degrees of translational and rotational freedom as well as the conformational degrees of freedom of both the ligand and protein, which provide the rigorous results for future analysis [20-26].
In this regard, in the present study, we are intended to identify the role of various flavanols as Cdk4 inhibitors. Flavanols are the category of chemical compounds, which majorly found in plant kingdom as a derivative of Flavans. The anticancer activities of these flavonoids are extensively studied but, till now no study has reported the effect of these flavanoids as inhibitor of Cdk4 for possible benefit in RB. Therefore, we have chosen certain flavanoids as given in Table 1 to perform docking with Cdk4 protein mode. The docking study has been carried out with the help of Molecular docking server by taking crystal structure of Cdk4 protein PDB ID: 2W9Z [27] from the www.pdb.org.
| S. No. | Name | Structure | Molecular formula |
|---|---|---|---|
| 1 | Catechin | C15H14O6 | 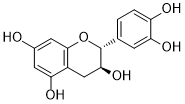 |
| 2. | Mesquitol | C15H14O6 | 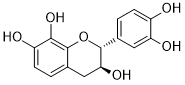 |
Table 1. Flavanoids to perform docking with Cdk4 protein mode.
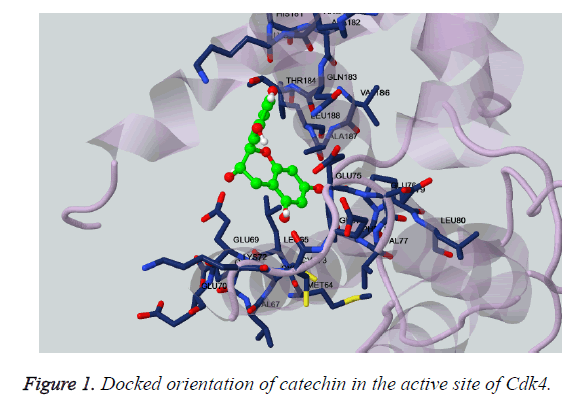
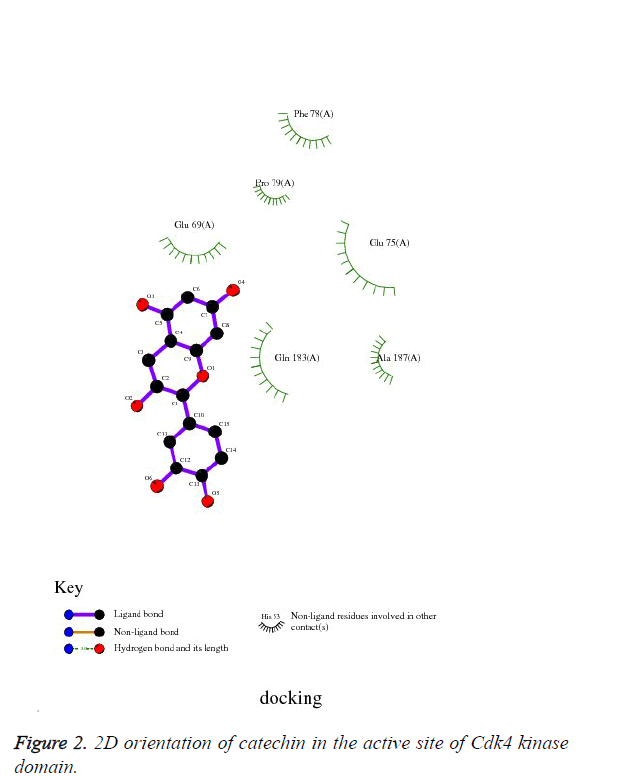
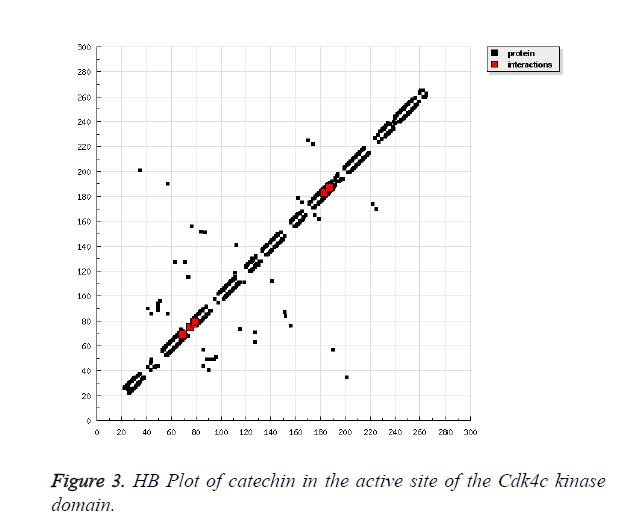
Catechins are the class of chemical constituents which are found in many of food and beverages. It considered as a type of phenol and medicinally acts as antioxidant [28-30]. Thus, its docking has been performed with Cdk4 kinase domain. As shown in the Figure 1, the catechin has been found highly oriented in the active site of the receptor Cdk4. The kinase domain of the Cdk4 has been shown in the typical bilobal structure for clear representation. It has been found that, the highly conserved residue of the ATP binding site of the protein were involved in making the prolific interaction with the catechin, As presented in the Table 2, the hydroxyl fragment of the ligand shown to engage the Glu69, Glu75 and Glu183 via the formation of three-polar bond. Whereas, the aromatic moiety of the Catechin shown to engage the Ala187 for hydrophobic interaction and Phe78 for cation-pi interaction. The other non-bonded interaction was also observed with the involvement of Glu69, Phe78, Pro79 and Gln183. The similar value of interaction has been also reported on close inspection of the HB plot of the catechin as shown in Figures 2 and 3.
| S. No | Name | Polar | Hydrophobic | Cation-pi | Other |
|---|---|---|---|---|---|
| 1 | Catechin | Glu69, Glu75, Glu183 |
Ala187 | Phe78 | Glu69, Phe78, Pro79, Gln183 |
| 2. | Mesquitol | Glu69, Glu75, Glu183 |
Cys68 | Phe78 | Cys68, Glu69, Phe78, Pro79, Lys180, Gln183, Thr184, Ala187 |
Table 2. Interaction table of studied flavanol with the Cdk4 kinase.
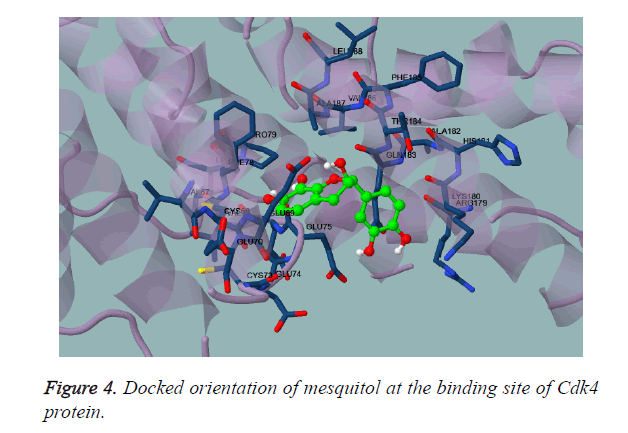
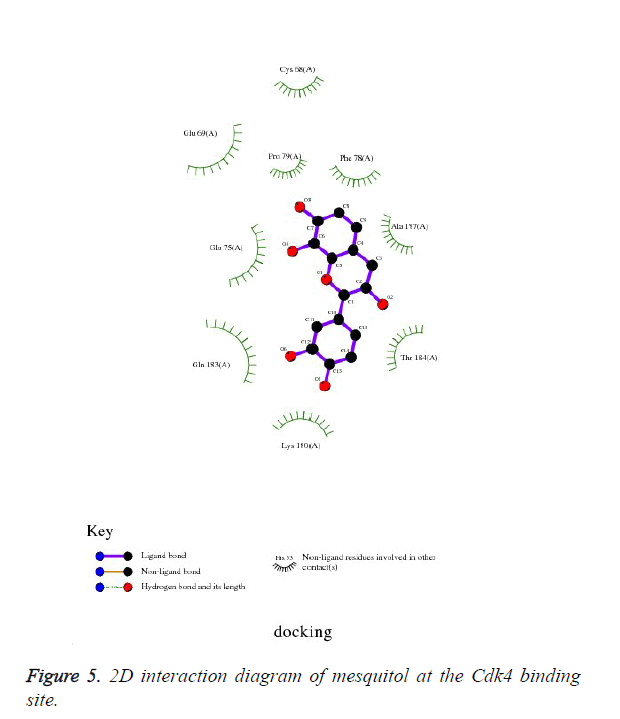
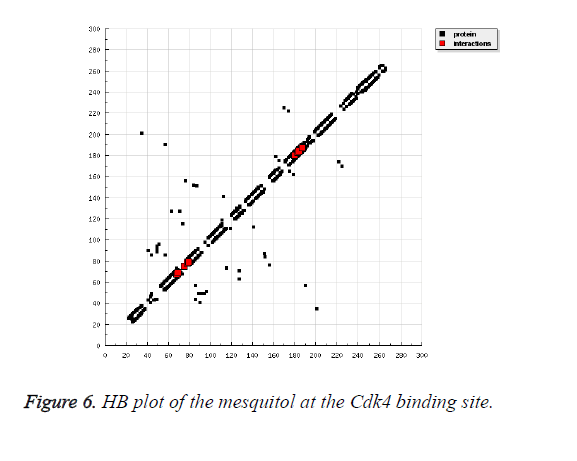
Mesquitol, is obtained from the Prosopis juliflora which chemical obtained as a (-) - mesquitol from the heartwood of the plant [31-33]. It also possesses antioxidant activity. Thus, on docking with ATP binding site of Cdk4 kinase, it shows very prolific engagements with the neighbouring residues as shown in Figure 4. As shown in Figure 5, the compound was found to be deeply buried in the active site of the ATP binding domain. The residues are highly innervated to take part in the binding as shown in the Figure 6, the 2D interaction profile of the mesquitol.
It has been found that, due to structural difference in the position of the hydroxyl group at the chromene nucleus, the mesquitol showed additional bonds with the neighbouring residues, such as, it showed to interact with, Cys68 for hydrophobic interaction and Gln183, Thr184 and Ala187 to create another non bonded interaction.
On closely inspecting the scoring profile of the ligands as presented in Table 3, it has been found that both the ligand create highly intriguing affinity for the receptor studies, i.e. Cdk4. In the case of catechin, the estimated free energy of the binding was revealed to -3.86 kcal/mol together with Ki at 1.49 mM. In regard of the above, the mesquitol showed very potent Ki of 52.95 μM accompanied with -5.83 μM. It confirmed that despite of having the basic chromene nucleus, both molecules exhibit diverse affinity towards the ATP binding site of the Cdk4. This could be easily interpreted on the basis of structural difference which makes mesquitol more potent than catechin.
| S. No. | Name | Estimated free energy of binding | Estimated inhibition constant, Ki | vdW+Hbond+desolv energy | Electrostatic energy | Total intermolecular energy |
|---|---|---|---|---|---|---|
| 1. | Catechin | -3.86 kcal/mol | 1.49 mM | -4.61 kcal/mol | -0.26 kcal/mol | -4.87 kcal/mol |
| 2. | Mesquitol | -5.83 kcal/mol | 52.95 μM | -5.88 kcal/mol | -0.44 kcal/mol | -6.32 kcal/mol |
Table 3. Scoring values of various flavanol as obtained by docking.
Conclusion
As a concluding remark, we have efficiently elucidated the effect of catechin and mesquitol on Cdk4 ATP binding site with the help of molecular docking. This study may open new avenue for the discovery of novel inhibitors targeting the retinoblastoma protein for the treatment of retinoblastoma.
Experimental
Docking calculations were carried out using docking server [34]. Gasteiger partial charges were added to the ligand atoms. Non-polar hydrogen atoms were merged, and rotatable bonds were defined. Docking calculations were carried out on Cdk-4 protein model. Essential hydrogen atoms, Kollman united atom type charges, and solvation parameters were added with the aid of Auto Dock tools [35]. Affinity (grid) maps of 60 × 60 × 60 Å grid points and 0.375 Å spacing were generated using the Autogrid program. Auto Dock parameter set- and distance-dependent dielectric functions were used in the calculation of the van der Waals and the electrostatic terms, respectively. The docking simulations were performed using the Lamarckian Genetic Algorithm (LGA) and the Solis and Wets local search method. Initial position, orientation, and torsions of the ligand molecules were set randomly. All rotatable torsions were released during docking. Each docking experiment was derived from 10 different runs that were set to terminate after a maximum of 250000 energy evaluations. The population size was set to 150. During the search, a translational step of 0.2 Å, and quaternion and torsion steps of 5 were applied.
References
- Popat K, McQueen K, Feeley TW. The global burden of cancer. Best Pract Res Clin Anaesthesiol 2013; 27: 399-408.
- Kanavos P. The rising burden of cancer in the developing world. Ann Oncol 2006;17: 14-23.
- Fitzmaurice C, Dicker D, Pain A, Hamavid H, Moradi-Lakeh M, MacIntyre MF. The global burden of cancer. JAMA Oncol 2015; 1: 505-527.
- Rodriguez-Galindo C, Orbach DB, VanderVeen D. Retinoblastoma. Paediatric Clinics N Am 2015; 201-23.
- Weinberg RA. The retinoblastoma protein and cell cycle control. Cell 1995; 323-330.
- Dimaras H, Kimani K, Dimba EAO, Gronsdahl P, White A, Chan HSL. Retinoblastoma. Lancet 2012; 379: 1436-1440.
- Aerts I, Lumbroso-Le Rouic L, Gauthier-Villars M, Brisse H, Doz F, Desjardins L. Retinoblastoma. Orphanet J Rare Dis 2006; 1: 31.
- Pandey AN. Retinoblastoma: An overview. Saudi J Ophthalmol 2014; 310-315.
- Ortiz MV, Dunkel IJ. Retinoblastoma. J Child Neurol 2016; 31: 227-236.
- Finger PT, Kurli M. Laser photocoagulation for radiation retinopathy after ophthalmic plaque radiation therapy. Br J Ophthalmol 2005; 89: 730-738.
- Wen JC, McCannel TA. Treatment of radiation retinopathy following plaque brachytherapy for choroidal melanoma. Curr Opin Ophthalmol 2009; 20: 200-244.
- Ng EYJ, Connolly BP, McNamara JA, Regillo CD, Vander JF, Tasman W. A comparison of laser photocoagulation with cryotherapy for threshold retinopathy of prematurity at 10 years: Part 1. Visual function and structural outcome. Ophthalmol 2002; 109: 928-934.
- Paysse EA, Lindsey JL, Coats DK, Contant CF, Steinkuller PG. Therapeutic outcomes of cryotherapy versus transpupillary diode laser photocoagulation for threshold retinopathy of prematurity. J Am Assoc Pediatr Ophthalmol Strabismus 1999; 3: 234-240.
- Saddic LA, West LE, Aslanian A, Yates JR, Rubin SM, Gozani O. Methylation of the retinoblastoma tumor suppressor by SMYD2. J Biol Chem 2010; 285: 37733-37740.
- Knudsen ES, Knudsen KE. Retinoblastoma tumour suppressor: where cancer meets the cell cycle. Exp Biol Med 2006; 231: 1271-1281.
- Sage J. The retinoblastoma tumour suppressor and stem cell biology. Genes Dev. 2012; 26: 1409-1420.
- Spring L, Bardia A, Modi S. Targeting the Cyclin D-Cyclin-Dependent Kinase (CDK) 4/6-retinoblastoma pathway with selective CDK 4/6 inhibitors in hormone receptor-positive breast cancer: rationale, current status, and future directions. Discov Med. 2016; 21: 65-74.
- Yu B, Lane ME, Pestell RG, Albanese C, Wadler S. Downregulation of cyclin D1 alters cdk 4- and cdk 2-specific phosphorylation of retinoblastoma protein. Mol cell Biol Res Commun 2000; 3: 352-359.
- Aron PM, Kennedy JA. Flavan-3-ols: Nature, occurrence and biological activity. Mol Nutr Food Res 2008; 52: 79-104.
- Meng XY, Zhang HX, Mezei M, Cui M. Molecular docking: A powerful approach for structure-based drug discovery. Curr Comput Aided Drug Des 2011; 7: 146-157.
- Kitchen DB, Decornez H, Furr JR, Bajorath J. Docking and scoring in virtual screening for drug discovery: Methods and applications. Nat Rev Drug Discov 2004; 3: 935-949.
- Shoichet BK, McGovern SL, Wei B, Irwin JJ. Lead discovery using molecular docking. Curr Opin Chem Biol 2002; 6: 439-446.
- Kitchen D, Decornez H, Furr J, Bajorath J. Docking and scoring in virtual screening for drug discovery: Methods and applications. Nat Rev Drug Discov 2004; 3: 935-949.
- Gschwend D, Good C, Kuntz ID. Molecular docking towards drug discovery. J Mol Recognit 1996; 9:175-186.
- Hirayama N. Docking method for drug discovery. Yakugaku Zasshi 2007; 127: 113-122.
- Ou Yang SS, Lu JY, Kong XQ, Liang ZJ, Luo C, Jiang H. Computational drug discovery. Acta Pharmacol Sin 2012; 33: 1131-1140.
- Protein data bank, 2016.
- Vuong QV, Golding JB, Nguyen M, Roach PD. Extraction and isolation of catechins from tea. J Separ Sci 2010; 33: 3415-3428.
- Yilmaz Y. Novel uses of catechins in foods. Tr Food Sci Technol 2006; 17: 64-71.
- Dong JJ, Ye JH, Lu JL, Zheng XQ, Liang YR. Isolation of antioxidant catechins from green tea and its decaffeination. Food Bioprod Process 2011; 89: 62-66.
- Sirmah P, Dumarcay S, Masson E, Gerardin P. Unusual amount of (-)-mesquitol from the heartwood of Prosopis juliflora. Nat Prod Res 2009; 23: 183-189.
- Jagadeeshwar RR, Tiwari AK, Kumar US, Reddy SV, Ali AZ, Rao JM. Novel 3-O-acyl mesquitol analogues as free-radical scavengers and enzyme inhibitors: Synthesis, biological evaluation and structure-activity relationship. Bioorganic Med Chem Lett 2003; 13: 2777-2780.
- Sirmah P, Mburu F, Iaych K, Dumarcay S, Gerardin P. Potential antioxidant compounds from different parts of Prosopis juliflora. J Trop For Sci 2011; 23: 187-195.
- Bikadi Z, Hazai E. Application of the PM6 semi-empirical method to modeling proteins enhances docking accuracy of AutoDock. J Cheminform 2009; 1: 15.
- Morris GM, Goodsell DS, Halliday RS, Huey R, Hart WE, Belew RK. Automated docking using a lamarckian genetic algorithm and an empirical binding free energy function. J Comput Chem 1998; 19: 1639-1662.