Review Article - Journal of Molecular Oncology Research (2018) Volume 2, Issue 3
Developments in the area of bladder cancer genomics and its importance in the treatment selection
Jyoti Sharma1,2, Barnali Deb1,2 and Prashanth Kumar1,2*
1Institute of Bioinformatics, International Technology Park, Bangalore, India
2Manipal Academy of Higher Education (MAHE), India
- *Corresponding Author:
- Prashanth Kumar
Institute of Bioinformatics, International Technology Park
Bangalore, India
E-mail: prashant@ibioinformatics.org
Accepted date: August 31, 2018
Citation: Sharma J, Deb B, Kumar P. Developments in the area of bladder cancer genomics and its importance in the treatment selection. J Mol Oncol Res. 2018;2(3):58-62.
DOI: 10.35841/molecular-oncology.2.3.58-62
Visit for more related articles at Journal of Molecular Oncology ResearchAbstract
With the advent of next-generation sequencing technologies, tremendous progress has been made in the understanding of the genomic landscape of several tumor types. Clinicopathologically bladder carcinoma is classified into non-muscle invasive bladder cancer and muscle invasive bladder cancer. Patients are often diagnosed with bladder carcinoma at an advanced stage, owing to a lack of early clinical symptoms and effective biomarkers for early detection. Failure of chemoradiotherapy at an advanced stage usually results in a poor outcome. Currently, there are limited targeted therapies available for bladder carcinoma. Thus, there is an immediate need to identify alternative strategies and novel therapeutic targets for the treatment of bladder carcinoma. The genomic underpinnings of bladder carcinoma may lead to the better understanding of this disease and improved targeted therapy. A large number of genomic alterations including somatic mutations, copy number alterations and fusion genes were identified to be involved in the pathogenesis of bladder carcinoma. A high mutational burden was observed in bladder carcinoma as compared to other tumors. Recurrent somatic mutations in genes such as TP53, KMT2D, RB1, FGFR3, KDM6A, STAG2 and PIK3CA were identified. Multiple signaling pathways such as the cell cycle pathway, DNA repair pathway, chromatin remodeling and histone modifications were found to be altered. Most likely, targeting mutated genes of these altered pathways could provide opportunities for personalization of bladder carcinoma therapy. This review discusses the molecular alterations, altered identified genes and the molecular pathways involved in bladder carcinoma.
Keywords
Urothelial carcinoma, NGS, Exome, Gene fusion, Alterd signaling pathways
Introduction
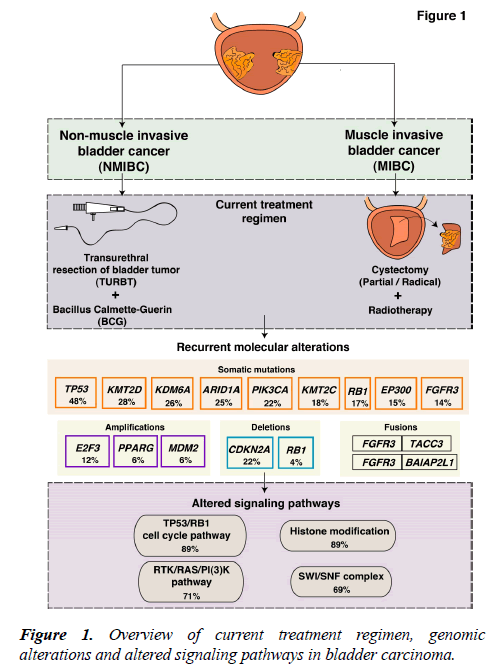
Bladder carcinoma is one of the most common malignancies of the urinary system with a high mutational load as compared to other solid tumors [1]. At diagnosis, the majority of bladder carcinoma cases (75%-80%) are non-muscle invasive bladder cancer (NMIBC) (Ta, non invasive papillary carcinoma; T1, tumor invades subepithelial connective tissue) [2] and 20%-25% of cases present as muscle invasive bladder carcinoma (MIBC) (T2-T4) [3,4]. NMIBC cases have a high recurrence rate (50%-70%) and moderate progression rate (10%-15%) [4]. NMIBC was initially suggested to arise from epithelial hyperplasia accounting for the loss of chromosome 9, which was considered an early event in tumorigenesis. Whereas, MIBCs were contemplated to be more genomically instable and arise mainly from dysplasia [5].
The current treatment regimens and interventions are mostly determined by the clinicopathological characteristics of the tumor. NMIBCs are generally treated with transurethral resection of bladder tumor (TURBT), by administration of Bacillus Calmette-Guerin (BCG) and intravesical chemotherapy [6]. The standard therapy for MIBC treatment is radical cystectomy (Figure 1). However, a perioperative mortality rate of 2.5%-5.2% exists [7,8]. In case of recurrence, patients are subjected to a numerous cystoscopic examinations. Thus, NMIBC is an expensive cancer to treat that place a huge demand on health care [9]. Systemic treatment for bladder carcinoma has been inadequate and limited to cisplatin-based chemotherapy with little progress over the past several decades. Furthermore, the development of multiple tumors in the same patient is conversant to the bladder carcinoma. A single clone may spread via intraepithelial migration to develop into more than one independent tumors [10-12]. This accounts for the intratumoral heterogeneity of these tumors and treatment becomes further challenging.
During the last decade, many research groups including the International Cancer Genome Consortium (ICGC), the Cancer Genome Atlas (TCGA), and the Chinese Cancer Genome Consortium (CCGC) conducted both large-scale and small-scale cancer genome studies worldwide. These studies identified molecular alterations including somatic mutations, copy number alterations and chromosomal rearrangements in various tumor types including bladder carcinoma. Bladder carcinoma has the third highest mutational frequency followed by lung cancer and melanoma [1]. Targeting common alterations in bladder carcinoma could result in novel trial design to determine the best therapy for patients. An overview of known molecular alterations in bladder carcinoma is shown in Figure 1. The targeted therapies approved by FDA for the treatment of bladder carcinoma are: Atezolizumab (Tecentriq™), nivolumab (Opdivo®), durvalumab (Imfinzi™), avelumab (Bavencio®), pembrolizumab (Keytruda®) (cancer.gov/about-cancer/treatment/types/targeted-therapies/ targeted-therapies-fact-sheet). Furthermore, longitudinal sequencing of matched primary and metastatic tissues from the same patients would strengthen the management of this disease. Recently, the introduction of immunotherapy in bladder carcinoma specifically the immune checkpoint inhibitors have been explored. Despite the successful introduction of various checkpoint inhibitors in bladder carcinoma, the objective response rate (ORR) lies within 46.4% ORR in programed death-ligand 1 (PD-L1) positive and between 0% and 26.2% ORR in PD-L1 negative patients [13-15]. Thus, most patients do not benefit from immunotherapy and hence the deeper understanding and detailed molecular characterization of bladder carcinoma is utmost needed.
Molecular alterations in bladder carcinoma
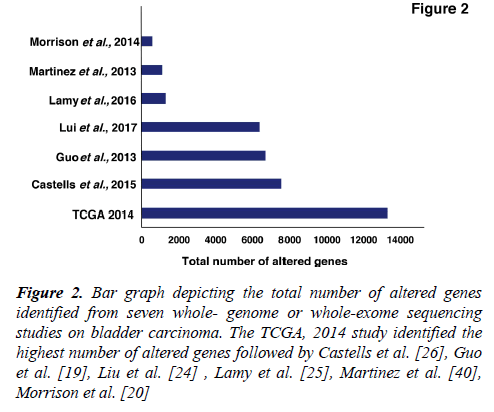
Several studies from various research groups including the TCGA group [16,17] and others [18-26] have carried out whole-genome sequencing (WGS) and whole-exome sequencing (WES) of bladder tumor tissue samples. A large number of genes were found to be mutated from different studies (Figure 2). In 2014, the TCGA group analyzed 131 high-grade MIBC cases and thirty-two significantly mutated genes were identified [16]. Recently, the TCGA extended the study and analyzed 412 MIBC cases. Fifty-eight significantly mutated genes were identified including TP53 (48%), KMT2D (28%), KDM6A (26%), ARID1A (25%), PI3K3CA (22%), KMT2C (18%), RB1 (17%), EP300 (15%), FGFR3 (14%), STAG2 (14%) [17]. These genes are known to play an important role in various functions, such as cell cycle regulation (TP53, RB1, CDKN1A and CDKN2A), chromosomal segregation (STAG2), chromatin remodeling (KDM6A, ARID1A, EP300, MLL2, BRWD1 and MBD1), receptor tyrosine kinase activity (FGFR3, TYRO3, ERBB3, TGFBR1, ERBB2, ERBB4 and IRS4), and migration of cells (RHOB and RHOA). Overall, one or more cell-cycle genes were altered in 93% of the TCGA tumor samples, most often TP53 and CDKN2A [16]. Interestingly, some of these mutations are mutually exclusive: CDKN2A and TP53, CDKN2A and RB1, TP53 and MDM2 [17], suggesting that they have redundant downstream targets affecting the pathogenesis of bladder carcinoma. The amplified/deleted genes were found to be involved in various important cellular processes including proliferation, apoptosis and migration of cells. A study by Guo et al. reported frequent alteration in STAG2, ESPL1 and NIPBL genes which are known to be involved in sister chromatid cohesion and segregation (SCCS) [19]. In contrast to other cancers, bladder cancer is a unique cancer type with genetic lesions in genes involved in the SCCS process. Cohesion is also affected by inactivation of the tumor suppressor pathway by inactivating RB1. This inactivation leads to defects in the SCCS process that further causes segregation errors in mitosis resulting in chromosomal instability. Lamy et al. performed WES of tumors from 29 patients initially diagnosed with early stage bladder tumors (14 with non-progressive disease and 15 with progressive disease). Tumors from patients with progressive disease showed a higher variance of the intrapatient mutational spectrum and a higher frequency of APOBEC-related mutations [25]. It has been reported by Glaser et al., that the APOBEC family of enzymes are major contributors to mutations and development of hypermutation phenotypes in bladder carcinoma [27]. The TCGA group determined that the high mutational load was associated with APOBEC-signature mutagenesis [17].
Figure 2: Bar graph depicting the total number of altered genes identified from seven whole- genome or whole-exome sequencing studies on bladder carcinoma. The TCGA, 2014 study identified the highest number of altered genes followed by Castells et al. [26], Guo et al. [19], Liu et al. [24] , Lamy et al. [25], Martinez et al. [40], Morrison et al. [20]
FGFR3 has long been implicated in bladder carcinoma. The mutations are known to affect kinase-activating sites and are amenable to therapeutic targeting [28]. A recurrent translocation FGFR3-TACC3 gene fusion on chromosome 4 (4p16.3) results in a constitutively activated FGFR3 receptor dimer. The breakpoints are located on exon 4 of TACC3 and on exon 18 of FGFR3 [29]. A total of nine FGFR3–TACC3 fusions and five FGFR3–BAIAP2L1 fusions were reported in bladder tumor cell lines and bladder tumors [30]. FGFR3 fusions were found to be extremely sensitive to FGFR-selective agents in the urothelial cell lines. The presence of a fusion gene(s) might aid in the selection of patients with bladder carcinoma for FGFR-targeted therapy [31]. A detailed list of frequently altered, amplified, deleted and fusion genes is provided in Table 1.
| Category | Altered genes | References |
|---|---|---|
| Frequently altered genes | TP53, CDKN1A, RB1, CDKN2A, STAG2, KDM6A, ARID1A, EP300, MLL2, BRWD1, MBD1, ERCC2, RHOB, RHOA, RHOA/RHOB, FAM47C, CHIT1, C3orf70, PIK3CA, TSC1, HRAS, KRAS, KRAS/HRAS, TXNIP, FGFR3, TYRO3, ERBB3, TGFBR1, ERBB2, ERBB4, IRS4, ELF3, ZFP36L1, RXRA, KLF5, NFE2L2, UTX, MLL3, CREBBP, NCOR1 | [16,17,19,23,24,32] |
| Amplified genes | BIRC3, BCL2L1, MDM2, CCNE1, CCND1, BEND3, E2F3, PVRL4, GDI2, PRKCI, FGFR3, ERBB2, EGFR, ZNF703, SOX4, PPARG, MYCL, MYC | [17,32] |
| Deleted genes | PDE4D, CDKN2A, RB1, ARID1A, CREBBP, NCOR1, FHIT, LRP1B, CCSER1, PTEN, FOXQ1, IKZF2 | [17,23,32] |
| Fusion genes | FGFR3–TACC3 and FGFR3–BAIAP2L1 | [16,19,30,31] |
| Altered signaling pathways | Cell cycle pathway, DNA repair pathway, RTK/RAS/PI(3) pathway, histone modifications and nucleosome complex pathway. | [16,17] |
Table 1: List of molecular alterations in bladder carcinoma.
Altered pathways and therapeutic targets of bladder carcinoma
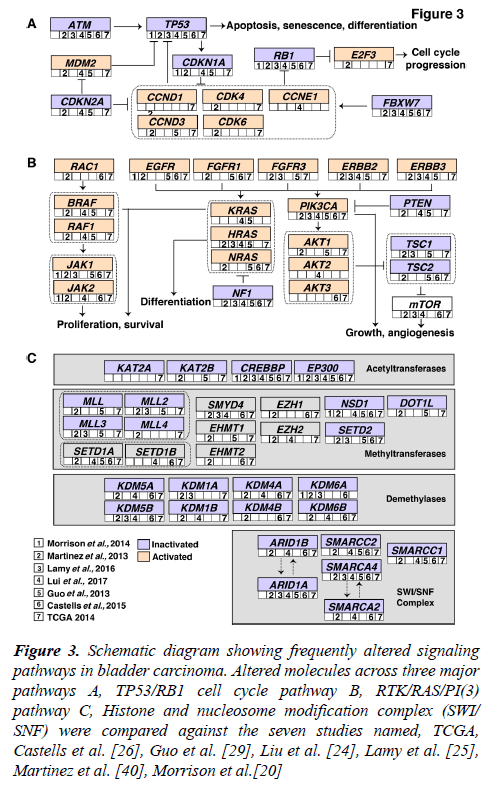
Accumulation of mutations result in acquired changes in the genome of patients with bladder carcinoma. These mutations have a major impact on the regulation of protein expression and function. The mutation frequency in NMIBC and MIBC overlap mutually. Large-scale genome-wide profiling poised to identify key signaling pathways altered in bladder carcinoma. These discovered pathways might be therapeutically targetable in the future. Thus, improving the prognosis of the bladder carcinoma patients. Frequent dysregulation of many key signaling pathways have been reported in bladder carcinoma by the TCGA group [16,17] (Figure 3). The cell cycle pathway is one of the major signaling pathway which has been reported to be altered in bladder carcinoma (Figure 3A). Inactivation of TP53, RB1 and CDKN2A was shown predominantly in MIBC [32,33]. CDKN1A, which is a cell cycle checkpoint regulating gene, is also found to harbor an inactivating mutation with a frequency of 11% [33]. TP53 and CDKN1A double loss of function mutants have been proposed to have enhanced sensitivity to CHK1 inhibitors where intervention with drugs such as Gemcitabine could result in better treatment [34]. Alterations in ERCC2 (9%) and ATM (14%) which were important DNA repair pathway intermediates and also observed to be altered in patients with bladder carcinoma. MDM2 amplification (6%) as well as overexpression (19%) has been observed in MIBC [17]. MDM2 is a direct transcriptional regulator of TP53 and it regulates an autoregulatory feedback loop. Moreover, MDM2 amplification and overexpression are reported to be mutually exclusive to TP53 mutations [17].
Figure 3: Schematic diagram showing frequently altered signaling pathways in bladder carcinoma. Altered molecules across three major pathways A, TP53/RB1 cell cycle pathway B, RTK/RAS/PI(3) pathway C, Histone and nucleosome modification complex (SWI/ SNF) were compared against the seven studies named, TCGA, Castells et al. [26], Guo et al. [29], Liu et al. [24], Lamy et al. [25], Martinez et al. [40], Morrison et al.[20]
The RTK/RAS/PI(3) pathway has been frequently observed in MIBCs (71%) to affect cell growth and proliferation [17]. The RTKs, EGFR, ERBB2, ERBB3, FGFR1 and FGFR3 are usually activated in bladder carcinoma which also activates RTK/RAS signaling. RAS–RAF signaling cascade further leads to phosphorylation of several downstream substrates that are responsible for multiple cellular effects such as proliferation and survival. Role of HRAS, NRAS, or KRAS in the RAS-RAF-MAPK pathway has been proven to control proliferation, differentiation, and survival of eukaryotic cells (Figure 3B) [35]. Activated RAS contributes to several phenotypic aspects of the malignant cells in bladder carcinoma including angiogenesis, cell growth and invasiveness [36,37]. Activated RAS may also directly activate PI3K. PIP3 recruits PDK1 and AKT, resulting in activation of AKT as well [4]. AKT1 is predominantly expressed in bladder carcinoma followed by AKT2 and AKT3. AKT1 is robustly involved in tumorigenesis and invasion of bladder carcinoma cells and it is evident that the inhibition of AKT1 could lead to suppressive effects in patients with bladder carcinoma [38]. EGFR mutations have been targeted by Erlotinib and Afatinib, and currently undergoing clinical trials in bladder carcinoma [37]. Similarly, ERBB2 amplifications are targeted by Trastuzumab and Pertuzumab; KRAS and NRAS by Cetuximab [37].
Pathways regulating the histone modification and chromatin remodeling (Figure 3C) are also frequently altered in bladder carcinoma. 89% of MIBCs harbor a mutation in one or more chromatin-regulating genes [16]. Several genes of the chromatin remodeling pathway such as ARID1A (4.2%), CREBB (14.2%) and KDM6A (4.9%) were deleted in bladder carcinoma. Also, histone methyltransferases including KMT2A, KMT2C and KMT2D, histone acetylases (CREBBP, EP300, KANSL1), ARID1A (SWI/SNF chromatin remodeling complex intermediate) and polycomb group genes (ASXL1, ASXL2) were observed to be frequently mutated [17]. Chromatin remodeling genes were frequently mutated in bladder carcinoma and targeting these genes would be advantageous for chromatin abnormalities. Drugs targeting histone modifying enzymes have received a wider recognition since such agents could effectively restore the epigenetic state of the genome back to normalcy due to the reversible histone modifications. Inhibitors targeting histone acetyltransferases, histone methyltransferases, histone demethylases are also moving rapidly to the clinical practice [39].
Conclusions
Bladder carcinoma is heterogeneous with a high genomic complexity. The large-scale sequencing efforts revealed signatures and underlying mutational spectrum. However, it is requisite to sequence large number of samples from patients with both NMIBC and MIBC to establish a more detailed mutational profile. Also, clinical trials are obligatory to stratify patients before treatment. We have provided a deeper insight into the genomic landscape of bladder carcinoma. The dysregulated pathways may be useful for designing targeted therapy to select the most effective treatment options for the intensive management of this disease.
Declarations
PK is a recipient of the Ramanujan Fellowship awarded by Department of Science and Technology (DST), Government of India. JS is a recipient of Bio-CARe Women Scientists award by Department of Biotechnology (DBT), Government of India. BD is a recipient of INSPIRE Fellowship from Department of Science and Technology (DST), Government of India.
References
- Alexandrov LB, Nik-Zainal S, Wedge DC, et al. Signatures of mutational processes in human cancer. Nature. 2013;500(7463):415-21.
- Brooks NA, O'Donnell MA. Treatment options in non-muscle-invasive bladder cancer after BCG failure. Indian J of Urol.2015;31(4):312-9.
- Ismaili N. Neoadjuvant Chemotherapy: A New Standard for Muscle Invasive Bladder Cancer? The Gulf J Oncolog. 2017;1(23):82-5.
- Knowles MA, Hurst CD. Molecular biology of bladder cancer: new insights into pathogenesis and clinical diversity. Nat Rev Cancer. 2015;15(1):25-41.
- Audenet F, Attalla K, Sfakianos JP. The evolution of bladder cancer genomics: What have we learned and how can we use it? Urol Oncol. 2018;36(7):313-20.
- Chang TC, Marcq G, Kiss B, et al. Image-Guided Transurethral Resection of Bladder Tumors - Current Practice and Future Outlooks. Bladder Cancer. 2017;3(3):149-59.
- Novara G, De Marco V, Aragona M, et al. Complications and mortality after radical cystectomy for bladder transitional cell cancer. J Urol. 2009;182(3):914-21.
- Stein JP, Lieskovsky G, Cote R, et al. Radical cystectomy in the treatment of invasive bladder cancer: long-term results in 1,054 patients. J Clin Oncol. 2001;19(3):666-75.
- Loras A, Trassierra M, Herraez SD, et al. Bladder cancer recurrence surveillance by urine metabolomics analysis. Sci Rep. 2018;8(1):9172.
- Simon R, Eltze E, Schafer KL, et al. Cytogenetic analysis of multifocal bladder cancer supports a monoclonal origin and intraepithelial spread of tumor cells. Cancer Res. 2001;61(1):355-62.
- Hafner C, Knuechel R, Zanardo L, et al. Evidence for oligoclonality and tumor spread by intraluminal seeding in multifocal urothelial carcinomas of the upper and lower urinary tract. Oncogene. 2001;20(35):4910-5.
- Cheng L, Gu J, Ulbright TM, et al. Precise microdissection of human bladder carcinomas reveals divergent tumor subclones in the same tumor. Cancer. 2002;94(1):104-10.
- Massard C, Gordon MS, Sharma S, et al. Safety and Efficacy of Durvalumab (MEDI4736), an Anti-Programmed Cell Death Ligand-1 Immune Checkpoint Inhibitor, in Patients With Advanced Urothelial Bladder Cancer. J Clin oncol. 2016;34(26):3119-25.
- Sharma P, Callahan MK, Bono P, et al. Nivolumab monotherapy in recurrent metastatic urothelial carcinoma (CheckMate 032): a multicentre, open-label, two-stage, multi-arm, phase 1/2 trial. Lancet Oncol. 2016;17(11):1590-8.
- Pichler R, Heidegger I, Fritz J, et al. PD-L1 expression in bladder cancer and metastasis and its influence on oncologic outcome after cystectomy. Oncotarget. 2017;8(40):66849-64.
- Comprehensive molecular characterization of urothelial bladder carcinoma. Nature. 2014;507(7492):315-22.
- Robertson AG, Kim J, Al-Ahmadie H, et al. Comprehensive Molecular Characterization of Muscle-Invasive Bladder Cancer. Cell. 2017;171(3):540-56.
- Yamashita M, Tsutsumi H, Sasaki M, et al. Halo-external fixation for a severe maxillofacial injury. Case report. Neurol Med Chir. 1985;25(6):484-8.
- Guo G, Sun X, Chen C, et al. Whole-genome and whole-exome sequencing of bladder cancer identifies frequent alterations in genes involved in sister chromatid cohesion and segregation. Nat Genet. 2013;45(12):1459-63.
- Pan H, Xu X, Wu D, et al. Novel somatic mutations identified by whole-exome sequencing in muscle-invasive transitional cell carcinoma of the bladder. Oncol Letters. 2016;11(2):1486-92.
- Zhao J, Xu W, He M, et al. Whole-exome sequencing of muscle-invasive bladder cancer identifies recurrent copy number variation in IPO11 and prognostic significance of importin-11 overexpression on poor survival. Oncotarget. 2016;7(46):75648-58.
- Morrison CD, Liu P, Read WA, et al. Whole-genome sequencing identifies genomic heterogeneity at a nucleotide and chromosomal level in bladder cancer. Proc Natl Acad Sci USA. 2014;111(6):E672-81.
- Nickerson ML, Dancik GM, Im KM, et al. Concurrent alterations in TERT, KDM6A, and the BRCA pathway in bladder cancer. Clin Cancer Research. 2014;20(18):4935-48.
- Liu D, Abbosh P, Keliher D, et al. Mutational patterns in chemotherapy resistant muscle-invasive bladder cancer. Nat Commun. 2017;8(1):2193.
- Lamy P, Nordentoft I, Demtroder BK, et al. Paired Exome Analysis Reveals Clonal Evolution and Potential Therapeutic Targets in Urothelial Carcinoma. Cancer Res. 2016;76(19):5894-906.
- Castells X, Karanovic S, Ardin M, et al. Low-Coverage Exome Sequencing Screen in Formalin-Fixed Paraffin-Embedded Tumors Reveals Evidence of Exposure to Carcinogenic Aristolochic Acid. Cancer Epidemiology Biomarkers Prev. 2015;24(12):1873-81.
- Glaser AP, Fantini D, Wang Y, et al. APOBEC-mediated mutagenesis in urothelial carcinoma is associated with improved survival, mutations in DNA damage response genes, and immune response. Oncotarget. 2018;9(4):4537-48.
- Cappellen D, De Oliveira C, Ricol D, et al. Frequent activating mutations of FGFR3 in human bladder and cervix carcinomas. Nat Genet. 1999;23(1):18-20.
- Scott SN, Ostrovnaya I, Lin CM, et al. Next-generation sequencing of urine specimens: A novel platform for genomic analysis in patients with non-muscle-invasive urothelial carcinoma treated with bacille Calmette-Guerin. Cancer Cytopathol. 2017;125(6):416-26.
- Martino DE, Tomlinson DC, Williams SV, et al. A place for precision medicine in bladder cancer: targeting the FGFRs. Future Oncol. 2016;12(19):2243-63.
- Costa R, Carneiro BA, Taxter T, et al. FGFR3-TACC3 fusion in solid tumors: mini review. Oncotarget. 2016;7(34):55924-38.
- Kim J, Akbani R, Creighton CJ, et al. Invasive Bladder Cancer: Genomic Insights and Therapeutic Promise. Clin Cancer Res. 2015;21(20):4514-24.
- Mitra AP, Datar RH, Cote RJ. Molecular pathways in invasive bladder cancer: new insights into mechanisms, progression, and target identification. J Clin Oncol. 2006;24(35):5552-64.
- Liu Y, Kwiatkowski DJ. Combined CDKN1A/TP53 mutation in bladder cancer is a therapeutic target. Mol Cancer Ther. 2015;14(1):174-82.
- Medarde FA, Santos E. Ras in cancer and developmental diseases. Genes Cancer. 2011;2(3):344-58.
- Ouerhani S, Elgaaied AB. The mutational spectrum of HRAS, KRAS, NRAS and FGFR3 genes in bladder cancer. Cancer Biomark. 2011;10(6):259-66.
- Felsenstein KM, Theodorescu D. Precision medicine for urothelial bladder cancer: update on tumour genomics and immunotherapy. Nat Rev Urol. 2018;15(2):92-111.
- Sabbineni H, Alwhaibi A, Goc A, et al. Genetic deletion and pharmacological inhibition of Akt1 isoform attenuates bladder cancer cell proliferation, motility and invasion. Eur J Pharmacol. 2015;764:208-14.
- Nair SS, Kumar R. Chromatin remodeling in cancer: a gateway to regulate gene transcription. Mol Oncol. 2012;6(6):611-9.
- Martínez BC, Pinilla RM, Casanova A, et al. ARID1A alterations are associated with FGFR3-wild type, poor-prognosis, urothelial bladder tumors. PLoS One. 2013;8(5):e62483.