Research Article - Biomedical Research (2017) Volume 28, Issue 5
Detection of HBV integration in plasma and paired tumor samples
Xiaofang Cui1*, Qing Huo2, Yanwei Qi2, Weiyang Li3*
1College of Life Sciences, Sichuan University, Cheng Du, PR China
2BGI-Shenzhen, Shenzhen, China
3Collaborative Innovation Center, Jining Medical University, Jining, PR China
- *Corresponding Author:
- Xiaofang Cui
College of Life Sciences, Sichuan University, PR China - Weiyang Li
Collaborative Innovation Center, Jining Medical University, PR China
Accepted date: October 31, 2016
Abstract
Hepatitis B viral (HBV) infection is one of the major causes of Hepatocellular Carcinoma (HCC). Previous studies had provided evidence that HBV integration may be an important factor for HCC carcinogenesis. In order to establish a new strategy for HCC diagnosis, a novel method was deployed to screen for HBV integration sites in both the HCC tumor and the paired plasma samples. HBV (type B) infections were detected in both patients, and a total number of 24 HBV viral integration sites were documented. Interestingly, one of the integration sites, in particular, was detected in both the HCC tumor and the paired plasma samples. In addition, it was noticed that HBV breakpoints inclined to the region of X protein (1,700-2,000 bp) in both the HCC tumor and the paired plasma samples. Altogether, our results provided evidence for HBV integration in plasma DNA, and they might be potentially useful for future HCC prognosis and diagnosis.
Keywords
Hepatocellular carcinoma, Plasma, HBV integration.
Introduction
HBV infection is a leading cause for chronic hepatitis, cirrhosis, and Hepatocellular Carcinoma (HCC) [1,2]. HBV carriers have high susceptibilities for cirrhosis and HCC [3]. HBV infection is an epidemic in Asia, Africa, Southern Europe and Latin America, and consists of at least eight genotypes (AH) [4]. In particular, HBV/A and HBV/D have a worldwide distribution, HBV/B and HBV/C are more restricted to east/ south-east Asia, HBV/E in west/central Africa, and HBV/F and HBV/H in indigenous populations of the Americas [5].
Massive Parallel Sequencing (MPS) technology has provided unprecedented means to study HBV integration globally. Despite several studies carried out by other groups, in which the authors investigated the effects of HBV integration [6], the genes preferentially integrated by HBV [7], and the viralhuman chimeric transcript which may predispose risks to the development and progression of liver cancer [8]. However, we noted that, the cancer tissues samples analysed in these studies were collected via invasive procedures. In order to facilitate the clinical utilization of HBV integration, we decided to investigate HBV integration in the DNA samples from the plasma and paired tumor samples.
We collected the HCC tumor and paired plasma samples from 2 HCC patients, and applied a High throughput Virus Integration Detection (HIVID) method to detect HBV integration sites [9]. In total, we detected 24 integration sites, and one of these sites was detected in both the tumor and the paired plasma samples. Furthermore, we discovered several novel genes which were integrated by HBV. Altogether, our results provided evidence for the HBV integration in the plasma DNA, which might be useful for HCC prognosis and diagnosis.
Materials and Methods
Sample collection
We obtained the plasma and tumor tissue samples of two HCC patients (001, 002) from the No.2 People’s Hospital, Chengdu, China, and both patients had been diagnosed with concurrent HBV infections (Table 1). Both patients had signed the written informed consent form, and the study had been approved by the Ethics Review Committee in the University of Sichuan.
| Sample-id (Patient) | Age | Gender | hbsag | Tumor-grade | Liver- pathology |
|---|---|---|---|---|---|
| 001 | 52 | Female | Positive | Moderatelydifferentiated | Cirrhotic |
| 002 | 24 | Male | Positive | Moderately differentiated | No-cirrhotic |
Table 1: Clinical information.
HBV fragments enrichment and sequencing
The construction of sequencing library strictly followed the standard instructions provided by Illumina. Genomic DNA samples were sheared into 150-200 bp DNA fragments using Covaris E-210 (Covaris, Inc., Woburn, MA). The sheared fragments were purified, and their ends were blunted, “A” tailed, and then ligated to adaptors. The sequencing libraries were quantified using Bio analyser 2100 (Agilent Technologies, Santa Clara, CA). The hybridization procedures were carried out following MyGenostics’s GenCap™ Target Enrichment Protocol (GenCap™ Enrichment, MyGenostics, USA). The sequencing libraries were hybridized with HBV probes at 65°C for 24 hours, and subsequently subjected to washes to remove unbound. The eluted fragments were amplified by 18 PCR cycles in order to generate the sequencing library. Upon the successful completion, each library was further quantified and preceded to 101 cycles of paired-end index sequencing in the Illumina HiSeq 2000 sequencer according to the manufacturer’s official instruction (Figure S1).
Breakpoints detection and annotation of HBV integration sites
Deploying an algorithm established by our team previously [9], the low-quality reads, duplication reads and also reads contain adaptor contaminations were removed. Subsequently, the filtered clean reads were mapped to both the human (NCBI build 37, HG19) and the HBV genomes. The chimeric reads (partially aligned to the human genome and partially aligned to the HBV genome were remained as the reads of our interest. The selected chimeric reads were then subjected to paired-end reads assembly, which helps to reconstruct fragment sequences, and additionally increase the efficacy to locate the precise position of the breakpoints. The PE-assembled reads were re-mapped to the human and the HBV genome using BWA (Figures S2 and S3) [10]. The HBV integration breakpoints were annotated using ANNOVAR [11].
Results
The HBV genotype and coverage of HBV genome
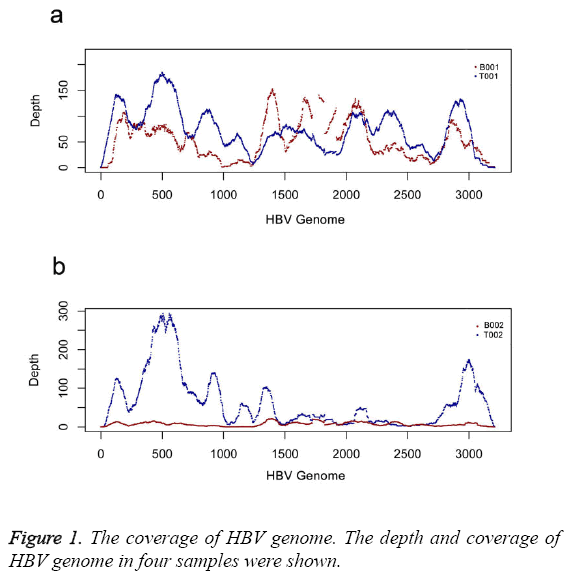
We calculated the coverage of two sets of samples. The results indicated two sets of samples were infected by HBV B type. The coverage of HBV genome was 98% (T001), 99% (B001), 100% (T002) and 99% (B002), respectively (Table 2). Depth distribution of two tumor samples shared the similar pattern (Figure 1). Two plasma samples had low depth distribution than that of tumor samples after depth was normalized (Figure 1). There was higher depth distribution in plasma sample of B001 than that in plasma sample of B002 (Figure 1).
| Tissue | Library | Total bases | Q20 | Effect reads | HBV type | HBV coverage | Breakpoint number |
|---|---|---|---|---|---|---|---|
| Plasma of patient 001 | B001 | 5.42G | 88.42;79.57 | 40162388(74.15%) | B | 99% | 3 |
| Plasma of patient 002 | B002 | 5.01G | 86.90;78.08 | 32108328(64.06%) | B | 98% | 4 |
| Tumor of patient 001 | T001 | 1.76G | 88.39;80.51 | 15056640(85.47%) | B | 100% | 15 |
| Tumor of patient 002 | T002 | 1.51G | 87.02;79.46 | 10358758(68.71%) | B | 99% | 2 |
Table 2: Data production of 4 tested samples.
Detection of HBV integration sites in tumor and paired plasma
Two sets of samples had been carried on detection of HBV integration by HIVID [9]. The results showed that there were 24 integration sites detected in both tumor and plasma samples. Among 24 breakpoints, 12 breakpoints were located in the genetic region and 12 breakpoints were located in the intergenic region. Breakpoint (chr18:11550976) could be detected in both T001 (tumor) and B001 (paired plasma) (Table 3). While there were no consistent integration sites between T002 and B002.
| Sample | Chr | Pos | Gene element | Gene | Total support reads |
|---|---|---|---|---|---|
| T002 | chr20 | 11684099 | Intergenic | LOC339593/BTBD3 | 4 |
| T002 | chr20 | 11683906 | Intergenic | LOC339593/BTBD3 | 31 |
| B002 | chr5 | 52674905 | Intergenic | LOC257396/FST | 2 |
| B002 | chr3 | 42691176 | Upstream | ZBTB47 | 2 |
| B002 | chr12 | 38638139 | Intergenic | ALG10B (Dist=72418) | 4 |
| B002 | chr2 | 28693259 | Intergenic | FOSL2/PLB1 | 2 |
| B001 | chr2 | 219543519 | Intronic | STK36 | 2 |
| B001 | chr2 | 219543501 | Intronic | STK36 | 4 |
| B001 | chr18 | 11550976 | Intergenic | PIEZO2/SLC35G4 | 11 |
| T001 | chr1 | 193627094 | Intergenic | CDC73 (Dist=403152) | 2 |
| T001 | chr6 | 135263065 | Intronic | ALDH8A1 | 3 |
| T001 | chr1 | 92114990 | Intergenic | CDC7/TGFBR3 | 3 |
| T001 | chr11 | 73919580 | Intronic | PPME1 | 2 |
| T001 | chr17 | 72816174 | Intronic | TMEM104 | 2 |
| T001 | chr14 | 72231097 | Intergenic | SIPA1L1/RGS6 | 2 |
| T001 | chr8 | 59068668 | Downstream | FAM110B | 4 |
| T001 | chr14 | 55444360 | Intronic | WDHD1 | 2 |
| T001 | chr6 | 42173478 | Downstream | MRPS10 | 2 |
| T001 | chr3 | 37934634 | Intronic | CTDSPL | 3 |
| T001 | chr19 | 36212332 | Exonic | KMT2B | 2 |
| T001 | chr17 | 18778353 | Intronic | PRPSAP2 | 10 |
| T001 | chr2 | 18395748 | Intergenic | KCNS3/NT5C1B-RDH14 | 2 |
| T001 | chr18 | 11550976 | Intergenic | PIEZO2/SLC35G4 | 71 |
| T001 | chr18 | 11550875 | Intergenic | PIEZO2/SLC35G4 | 2 |
Table 3: The position of breakpoints.
The distribution of HBV breakpoints in HBV genome
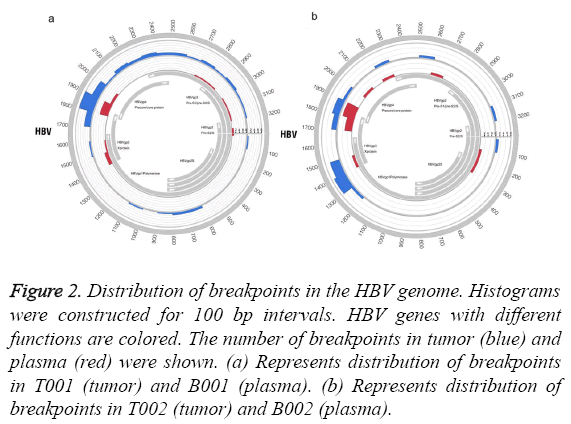
We analysed the distribution of HBV breakpoints in the HBV genome. The results revealed that samples of tumor and plasma shared the similar hotspot region of HBV integration (Figure 2). HBV integration was inclined to occur in the region (1700-2000 bp) of the HBV genome (Figure 2).
Figure 2: Distribution of breakpoints in the HBV genome. Histograms were constructed for 100 bp intervals. HBV genes with different functions are colored. The number of breakpoints in tumor (blue) and plasma (red) were shown. (a) Represents distribution of breakpoints in T001 (tumor) and B001 (plasma). (b) Represents distribution of breakpoints in T002 (tumor) and B002 (plasma).
Discussion
HBV integration had been shown linking to the tumorigenesis of HCC. The studies led by Sung et al. had identified several genes preferentially integrated by HBV [7]. In this study, we analysed the breakpoints of HBV integration in two sets of the paired plasma and the HCC tumor samples. In the first sample set, the same breakpoint could be detected in the HCC tumor (T001) tissue and the paired plasma (B001) samples. However, there were no identical integration sites found in the other set of samples. The use of plasma DNA sample is crucial for clinical diagnosis due to a certain percentage of circulating DNA originated from the degenerating tumor cells [12]. According to previous study, apoptosis might be one of the major sources of plasma or serum DNA [13], despite the entire mechanism of DNA being released into circulating blood still remain to be thoroughly investigated. Accordingly, plasma DNA had been proposed for early diagnosis [14]. The reason why there was no consistent integration site in the second set of the HCC tumor and the paired plasma samples is unclear. Curiously, we noted that the two patients recruited in this study had different clinical background (Table 1), which might indicate clinical factors had some effect on HBV integration in these samples. Owing to a limited number of patients, further studies probably are required to clarify this matter.
Furthermore, our findings also included several novel HBVintegrated genes. Among these genes, STK36, PPME1 were in particularly interesting, because of their associations with cancers. STK36 encodes a serine/threonine kinase, which is an established therapeutic target for cancer treatments [15]. PPME1 produces a protein phosphatase methyl esterase that has roles in malignant glioma progression [16].
Moreover, twelve breakpoints had been located to the intergenic regions. Nevertheless, the importance and significance of these integration sites remained elusive. Worthwhile to point out though, an increasing trend of research interests had drawn to resolve the usefulness of intergenic integration sites. For instances, MYC activation was driven by an upstream integration of HPV-18 genome [17]; β-catenin transactivity could be modulated by HBV integration in Long Interspersed Nuclear Element (LINE) [8].
The distribution of breakpoints in the HBV genome was also investigated. The breakpoints were particularly enriched in the coding regions of HBV X and core genes; this is consistent with the previous findings by others and also our group. The different depth pattern between the tumor and the plasma samples might be due to that the HBV DNA in the tumor and plasma samples might have different source.
Our study had provided evidence for HBV integration in plasma DNA, which might be potentially useful for future HCC prognosis and diagnosis.
Conflict of Interest Statement
The authors declare no competing interests.
Author's Contributions
Conceived and designed the experiments: CXF. Performed the experiments: CXF, WYL, and YWQ. Analysed the data: CXF, WYL. Wrote paper: HQ, CXF, and WYL.
References
- Ott JJ, Stevens GA, Groeger J, Wiersma ST. Global epidemiology of hepatitis B virus infection: new estimates of age-specific HBsAg seroprevalence and endemicity. Vaccine 2012; 30: 2212-2219.
- Murakami Y, Minami M, Daimon Y, Okanoue T. Hepatitis B virus DNA in liver, serum, and peripheral blood mononuclear cells after the clearance of serum hepatitis B virus surface antigen. J Med Virol 2004; 72: 203-214.
- Beasley RP. Hepatitis B virus. The major etiology of hepatocellular carcinoma. Cancer 1988; 61: 1942-1956.
- Lin CL, Liao LY, Wang CS, Chen PJ, Lai MY, Chen DS, Kao JH. Basal core-promoter mutant of hepatitis B virus and progression of liver disease in hepatitis B e antigen-negative chronic hepatitis B. Liver Int Offic J Int Assoc Study Liver 2005; 25: 564-570.
- Araujo NM, Waizbort R, Kay A. Hepatitis B virus infection from an evolutionary point of view: how viral, host, and environmental factors shape genotypes and subgenotypes. Infection, genetics and evolution. J Mol Epidemiol Evol Gene Infect Dis 2011; 11: 1199-1207.
- Jiang Z, Jhunjhunwala S, Liu J, Haverty PM, Kennemer MI, Guan Y, Lee W, Carnevali P, Stinson J, Johnson S. The effects of hepatitis B virus integration into the genomes of hepatocellular carcinoma patients. Genome Res 2012; 22: 593-601.
- Sung WK, Zheng H, Li S, Chen R, Liu X. Genome-wide survey of recurrent HBV integration in hepatocellular carcinoma. Nat Genet 2012; 44: 765-769.
- Lau CC, Sun T, Ching AK, He M, Li JW, Wong AM, Co NN, Chan AW, Li PS, Lung RW. Viral-human chimeric transcript predisposes risk to liver cancer development and progression. Cancer Cell 2014; 25: 335-349.
- Li W, Zeng X, Lee NP, Liu X, Chen S. HIVID: an efficient method to detect HBV integration using low coverage sequencing. Genomics 2013; 102: 338-344.
- Li H, Durbin R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics 2009; 25: 1754-1760.
- Wang K, Li M, Hakonarson H. ANNOVAR: functional annotation of genetic variants from high-throughput sequencing data. Nucleic Acids Res 2010; 38: e164.
- Anker P, Mulcahy H, Chen XQ, Stroun M. Detection of circulating tumour DNA in the blood (plasma/serum) of cancer patients. Cancer Metast Rev 1999; 18: 65-73.
- Elshimali YI, Khaddour H, Sarkissyan M, Wu Y, Vadgama JV. The clinical utilization of circulating cell free DNA (CCFDNA) in blood of cancer patients. Int J Mol Sci 2013; 14: 18925-18958.
- Jahr S, Hentze H, Englisch S, Hardt D, Fackelmayer FO, Hesch RD, Knippers R. DNA fragments in the blood plasma of cancer patients: quantitations and evidence for their origin from apoptotic and necrotic cells. Cancer Res 2001; 61: 1659-1665.
- Murone M, Luoh SM, Stone D, Li W, Gurney A. Gli regulation by the opposing activities of fused and suppressor of fused. Nat Cell Biol 2000; 2: 310-312.
- Puustinen P, Junttila MR, Vanhatupa S, Sablina AA, Hector ME, Teittinen K, Raheem O, Ketola K, Lin S, Kast J. PME-1 protects extracellular signal-regulated kinase pathway activity from protein phosphatase 2A-mediated inactivation in human malignant glioma. Cancer Res 2009; 69: 2870-2877.
- Adey A, Burton JN, Kitzman JO, Hiatt JB, Lewis AP. The haplotype-resolved genome and epigenome of the aneuploid HeLa cancer cell line. Nature 2013; 500: 207-211.