Research Article - Current Pediatric Research (2017) Volume 21, Issue 1
Copy number variations in chromosome 16p13.11-The neurodevelopmental clinical spectrum
Susana Loureiro1,2,3, Joana Almeida1, Cátia Café1, Inês Conceição4, Susana Mouga1, Ana Beleza5, Bárbara Oliveira4, Joaquim de Sá5, Isabel Carreira6, Jorge Saraiva2,5, Astrid Vicente4, Guiomar Oliveira1,2
1Neurodevelopment and Autism Unit (UNDA), Serviço do Centro de Desenvolvimento da Criança, Centro de Investigação e Formação Clínica, Hospital Pediátrico, Centro Hospitalar e Universitário de Coimbra, Coimbra, Portugal.
2University Clinic of Paediatrics, Faculty of Medicine, University of Coimbra, Coimbra, Portugal.
3Department of Paediatrics, Centro Hospitalar Tondela-Viseu, Viseu, Portugal.
4Instituto Nacional de Saúde Dr. Ricardo Jorge, Lisboa, Portugal; Instituto Gulbenkian de Ciência, Oeiras, Portugal; Center for Biodiversity, Functional and Integrative Genomics, Faculty of Science, University of Lisbon, Lisboa, Portugal
5Medical Genetics Unit, Hospital Pediátrico, Centro Hospitalar e Universitário de Coimbra, Coimbra, Portugal.
6Cytogenetics and Genomics Laboratory, Faculty of Medicine, University of Coimbra, Coimbra, Portugal.
- *Corresponding Author:
- Susana Loureiro
Pediatrician, Centro Hospitalar de Tondela-Viseu
Pediatrics, Viseu, Portugal.
Tel: 00351967420040
E-mail: susana_loureiro@hotmail.com
Accepted date: January 30, 2017
Abstract
Generalized clinical use of Chromosomal Microarray Analysis (CMA) in etiological investigation of neurodevelopmental disorders, has led to the decipherment of many new copy number variations (CNV), such as those in 16p13.11. We report detailed phenotypic and molecular characterization of four patients with duplications and four patients with deletions in 16p13.11.We found partially overlapping CNVs of different sizes with varied clinical presentation. Patients presented with Intellectual Disability (ID), Autism Spectrum Disorder (ASD) and behavioral abnormalities. Dysmorphisms and skeletal manifestations were seen in patients with duplications. Our report further delineates the phenotypic spectrum associated with 16p13.11 CNVs. The genes implicated in the region and their potential contribution to the phenotype is discussed with special attention to the yet not fully known C16orf45.
Keywords
Autism spectrum disorder, Intellectual disability, Chromosomal microarray analysis, DNA copy number variants, Chromosome 16p13.11.
Abbreviations
aCGH: Comparative Genome Hybridization Array; ADHD: Attention Deficit Hyperactivity Disorder; AGP: Autism Genome Project; ASD: Autism Spectrum Disorder; CMA: Chromosomal Microarray Analysis; CNV: Copy Number Variations; DQ: Development Quotient; GMDS: Griffiths Mental Development Scale; GNDD: Global Neurodevelopmental Delay; Hg: Human Genome; ID: Intellectual Disability; IQ: Intellectual Quotient; SNP: Single-Nucleotide Polymorphism Array; UNDA: Neurodevelopment and Autism Unit; VABS: Vineland Adaptive Behavior Scale; WISC-III: Wechsler Intelligence Scale for Children.
Introduction
The neurodevelopmental disorders are a group of conditions with onset in the developmental period. The neurodevelopmental disorders frequently co-occur; for example, individuals with autism spectrum disorder (ASD) often have intellectual disability (ID) [1].
ASD is characterized by persistent deficits in social communication and social interaction across multiple contexts. In addition, the diagnosis of ASD requires the presence of restricted, repetitive patterns of behavior, interests, or activities [1]. ASD is currently estimated to have a prevalence between 1% and 2% [1,2]. Overall estimated ASD prevalence in the United States was 11.3 per 1,000 (one in 88) in 2008 with a male/female ratio of 4.6 [3]. In Portugal, according to a study targeting children with six to nine years of age, in the school-year 1999-2000, the global prevalence of autistic disorder was approximately 10 per 10000 [4].
Autistic behavior is very frequent in individuals with a primary diagnosis of ID (rates from three to four times higher than in the general population), a genetically heterogeneous condition affecting 1-3% of the population [1,5]. Conversely, approximately 38%-83% patients with ASD have ID [4]. Consequently, it is widely believed that ID and autism are complex, multifactorial neurodevelopmental disorders with high heritability, which share not only some symptoms, but also overlapping risk factors [6,7].
Great progress has been made in the past years in identifying rare variants of major effect in both ASD and ID. However the genetic factors underlying these disorders remains mostly elusive [7]. Dozens of individually rare gene variants and loci associated with high-risk for ASD have been identified, which overlap extensively with genes for ID [7]. These findings, while demonstrating the heterogeneous etiology, provide important clues about the pathophysiology [8]. Estimates from ongoing studies estimate that 60-80% of ASD and ID-related genetic variants remain to be discovered [5,7,8]. Chromosomal microarray analysis (CMA), such as comparative genome hybridization (aCGH) and single-nucleotide polymorphism (SNP) arrays, recently expanded the range of studies at the molecular level, improving the ability to detect microdeletions and microduplications known as copy number variants (CNV) [9-13]. CNV analysis now offers researchers the first truly cost-effective tool for scanning the entire genome for rare variants [9,14,15].
Several recent CNV studies have identified specific regions of the genome that appear to carry substantial risk for some neurodevelopmental and neuropsychiatric disorders including ASD, ID, attention deficit hyperactivity disorder (ADHD) and schizophrenia [9-11,13,16]. CNVs are known to account for 5-10% of “idiopathic ASD” cases (those with no obvious clinical syndrome after a comprehensive clinical examination and laboratorial investigation) [8,10- 12,17,18]. With exome and whole-genome sequencing, in recent years, studies have estimated at least another 6% contribution to ASDand an additional 5% afforded by rare inherited recessive or X-linked loss-of-function singlenucleotide variations [19-25].
Assessing the clinical significance of rare CNVs is not straightforward, particularly due to the small number of reported cases, the marked variable expressivity and the identification of similar or identical rearrangements in apparently normal individuals, which may indicate either an incomplete penetrance or the benignity of the variant. Among these are the deletions and duplications in chromosome 16p13.11, which are not thoroughly characterized phenotypically [26-28].
Chromosome 16 is especially rich in intrachromosomal segmental duplications or low copy repeats (LCRs) that mediate recurrent genomic rearrangements, specially by non-allelic homologous recombination (NAHR) [26,27,29-31]. There is a widespread spectrum of phenotypes associated with CNVs of 16p13.11 [26-28]. Duplications in 16p13.11 region, initially considered to be rare benign variants, have been recently implicated in multiple neurodevelopmental, behavioral and physical abnormalities such as ASD, ID, schizophrenia, ADHD, other behavioral abnormalities and with cardiac and skeletal manifestations like hypermobility, craniosynostosis and polydactyly [16,26-28,31-37]. These duplications exhibit incomplete penetrance [16,26]. Deletions have been associated with focal and generalized epilepsy, multiple congenital anomalies, developmental delay/ID, microcephaly and ASD [26-28,31,38-44]. 16p13.11 CNVs are also linked to juvenile and childhood onset psychosis and obsessive-compulsive disorder [45,46].
We examined the clinical phenotype and genotype of eight patients with 16p13.11 CNVs in a cohort of the Neurodevelopment and Autism Unit (UNDA) of the Child Development Center at Pediatric Hospital of CHUC – Coimbra Hospital and University Centre.
Materials and Methods
Subjects
For the identification of the cases carrying CNVs in 16p13.11 region, we searched in the UNDA Database (clinical database of this unit).
At the time of analysis (August 2013), this clinical database contained 2818 clinical referrals, including 1708 patients referred for ASD (81% of whom were males), 377 with ID/global neurodevelopmental delay (GNDD), 61 with ADHD/behavioral problems, 140 with learning disabilities and 532 other diagnosis.
The clinical diagnosis was based on a multidisciplinary assessment by a clinical team with extensive experience in neurodevelopmental disorders (Table 1).
Neurodevelopmental disorders were defined according to the DSM-IV-TR (Diagnostic and Statistical Manual of Mental Disorders, fourth edition, Text Revision) criteria [47].
UNDA protocol - a panel of clinical diagnostic instruments - was used for assessing ASD. Children were diagnosed with ASD if met the cut-off for ASD on the Autism Diagnostic Interview-Revised (ADI-R), on the Autism Diagnostic Observation Schedule (ADOS), on the Childhood Autism Rating Scale (CARS) and fulfilled the DSM-IV-TR criteria for ASD [47-50].
The development quotient (DQ) was obtained by Griffiths Mental Development Scale (GMDS), the intellectual quotient (IQ) was evaluated using the Wechsler Intelligence Scale for Children-WISC-III and the adaptive functioning with the Vineland Adaptive Behavior Scale – VABS [51-53]. It was defined a mean score of 100 with a standard deviation of 15 for the GMDS, WISC III and VABS scales.
Patients with clinical criteria (ASD, ID, GNDD) underwent etiological investigation, most of them before the advent of the CMA. The initial genetic testing included karyotype, fragile-X analysis, subtelomeric rearrangements and analysis of 15p11-13, 16p11.2 and 22q13 regions. Patients underwent metabolic testing, if the former were negative, that included aminotransferases, ammonia, lactate, creatine phosphokinase blood levels and analysis of amino acids, organic acids, creatine metabolism, and CDT-carbohydrate deficient transferrin. In selected cases, other specific metabolic testing was performed, like acylcarnitine profile, long chain fatty acids and lysosomal enzymes. Head magnetic resonance imaging with spectroscopy and electroencephalogram was reserved for cases with neurological signs/symptoms.
| Diagnosis | Criteria |
|---|---|
| GNDD | In children aged less than six years if they perform more than two standard deviations (SDs) below age-matched peers in two or more aspects of development |
| ID | If significant limitations in both intellectual functioning and adaptive behavior were present and begun before the age of 18 years |
| Mild ID | If intellectual quotient was 50-70 |
| Moderate ID | If intellectual quotient was 35-49 |
| Severe ID | If intellectual quotient was <35 |
| ASD | UNDA Protocol: If fulfilled the cut-off for ASD on ADI-R ADOS CARS DSM-IV-TR criteria |
| Neurodevelopmental comorbidities* | DSM-IV-TR criteria |
| Epilepsy** | Condition characterized by recurrent (two or more) epileptic seizures, unprovoked by any immediate identified cause. Seizures were classified according to the initial semiology: focal if was consistent with initial activation of only part of one hemisphere and generalized if was consistent with more than minimal involvement of both cerebral hemispheres. |
| Abbreviations: GNDD: Global Neurodevelopmental Delay (Petersen et al [61], Michelson et al [15]); ID: Intellectual Disability (American Psychiatric Association [47]); ASD: Autism Spectrum Disorder; UNDA: Neurodevelopment and Autism Unit; ADI-R: Autism Diagnostic Interview-Revised (Lord et al. [48]); ADOS: Autism Diagnostic Observation Schedule (Lord et al. [49]); CARS: Childhood Autism Rating Scale (Shopler et al. [50]); DSM-IV-TR: Diagnostic and Statistical Manual of Mental Disorders, fourth edition, Text Revision (American Psychiatric Association [47]). *Other mental disorders like attention deficit hyperactivity disorder (ADHD), depressive disorder and obsessive-compulsive disorder (OCD); **(Blume et al [68]) | |
Table 1. Neurodevelopmental diagnosis criteria
CMA, either aCGH or SNP-arrays, was performed following a normal initial investigation or as a first-line test in 379 selected patients with idiopathic ID/GNDD or ASD. CNVs of clinical relevance (pathogenic CNVs) were detected in 39 patients (10.3%), eight of them being 16p13.11 CNVs (20.5% of clinically relevant CNVs and 2.1% of cases who performed a CMA). No other pathogenic CNVs were found.
The genomic DNA analysis of the patients were requested by Genetic Consultation or were identified by the SNP genotyping carried out by the Autism Genome Project (AGP), the world's largest research project on identifying genes associated with risk for ASD. The project is a research partnership involving approximately 50 academic and research institutions, including cases followed at UNDA, that have pooled DNA samples in a collaborative effort.
We obtained informed consent for investigation and publication in all cases.
CNV Identification
The configuration and resolution of the genomic DNA analysis and the software used for analysis of the result is summarized in Table 2.
The genomic coordinates were based on the NCBI Build 36.1 (UCSC Human genome-hg18, March 2006) [54].
CNV Validation
All de CNVs were validated by a second method. Three of our patients were analyzed by aCGH for etiological diagnosis purposes in a clinical setting. In two of them was used the Agilent 60mer oligonucleotide-array, configuration 4x180K with an average genome wide resolution of 60 Kb. Analysis of the results was made by Agilent Genomic Workbench Lite Edition 6.5. CMA was controlled using a loop design in which three hybridizations were carried out with three test patients that were compared with each other. In one patient was used the Human Genome CGH Microarray Kit 244A (Agilent), with an average genome wide resolution of 9 kb. After hybridization the array was scanned with the Agilent Microarray Scanner (G2565BA), and the results were analyzed using Feature Extraction Software v.9.1 and Genomic Worchbench 5.0 (Agilent; Parameter: Aberration Algorithm ADM-2; Threshold 5.9; 5 points aberration filter).
| Requesting of CMA | CMA | Analysis of the results | |
|---|---|---|---|
| Patient 1 | Genetic consultation | Agilent 60mer oligonucleotide-array; Configuration 4x180K; Average genome wide resolution 60 Kb | Agilent Genomic Workbench Lite Edition 6.5. |
| Patient 2 | Research study - Whole genome SNP array (Autism Genome Project [17,19]) | Illumina 1M Duo SNP microarray | Illumina Genome Viewer software using QuantiSNP, iPattern and PennCNV algorithms |
| Patient 3 | Genetic consultation | Human Genome CGH Microarray Kit 244A (Agilent); Average genome wide resolution 9 kb | Feature Extraction Software v.9.1 and Genomic Workbench 5.0. |
| Patient 4 | Research study - Whole genome SNP array (Autism Genome Project [17,19]) | Illumina Infinium 1M Single SNP microarray | Illumina Genome Viewer software using QuantiSNP, iPattern and PennCNV algorithms |
| Patient 5 | Genetic consultation | Agilent 60mer oligonucleotide-array; Configuration 4x180K; Average genome wide resolution 60 Kb | Agilent Genomic Workbench Lite Edition 6.5. |
| Patient 6 | Research study - Whole genome SNP array (Autism Genome Project [17,19]) | Illumina 1M Duo SNP microarray | Illumina Genome Viewer software using QuantiSNP, iPattern and PennCNV algorithms |
| Patient 7 | Research study - Whole genome SNP array (Autism Genome Project [17,19]) | Illumina Infinium 1M Single SNP microarray | Illumina Genome Viewer software using QuantiSNP, iPattern and PennCNV algorithms |
| Patient 8 | Research study - Whole genome SNP array (Autism Genome Project [17,19]) | Illumina Infinium 1M Single SNP microarray | Illumina Genome Viewer software using QuantiSNP, iPattern and PennCNV algorithms |
| Abbreviations: CMA: Chromosomal Microarray Analysis; Kb: Kilobase Pairs; 1M: 1 Million; SNP: Single-Nucleotide Polymorphism Array | |||
Table 2. Microarray analysis, configuration and resolution and software used for analysis of the results
The other five patients were genotyped in the context of SNP genotyping performed by the AGP using the Illumina 1M Duo SNP microarray platform or the Illumina Infinium 1M Single SNP microarray platform [19]. Rare de novo CNVs, clinically relevant CNVs, and other selected rare CNVs were validated by at least one method (quantitative Polymerase Chain Reaction - qPCR, MLPA- Multiplex ligation-dependent probe amplification, and/or longrange PCR) [19]. 16p13.11 CNVs validation by real time qPCR used the Universal Probe Library System (UPL) system and the LightCycler 480 from Roche. The UPL system consists of a probe-based detection protocol with a labelled hydrolysis probe that will anneal between the primer sites and consequently, will only generate a signal when the correct amplicon is produced. The quantitative PCR (qPCR) was performed using 25 ng of the genomic DNA template, two primer pairs specific for NTAN1, C16orf45 and ABCC1 genes, and the specific fluorescent probe for each amplicon. qPCR was carried out using standard conditions. Each assay was designed using the ProbeFinder Software available online at the Assay Design Center.
Despite the platform heterogeneity in CNV detection, numerous studies have shown that, whereas for small variations the CNV detection power is dependent on the array used, different array platforms have comparable sensitivity and specificity for the detection of large CNVs (>500 kbp in size) [29]. Since that the different array platforms used have an adequate probe coverage for the 16p13.11 region, platform-specific differences in detection are unlikely to represent a major confounding factor in our analyses.
Case Reports
Eight patients had 16p13.11 CNVs. Two had a diagnosis of GNDD/ID (patients 1 and 8) and six had ASD (patients 2-7) with or without ID.
Tables 3 to 6 summarize the clinical and molecular characterization of the eight patients with 16p13.11CNVs. The initial etiological investigation performed in patients 1-7 was negative. Patient 8 realized CMA as a first line test.
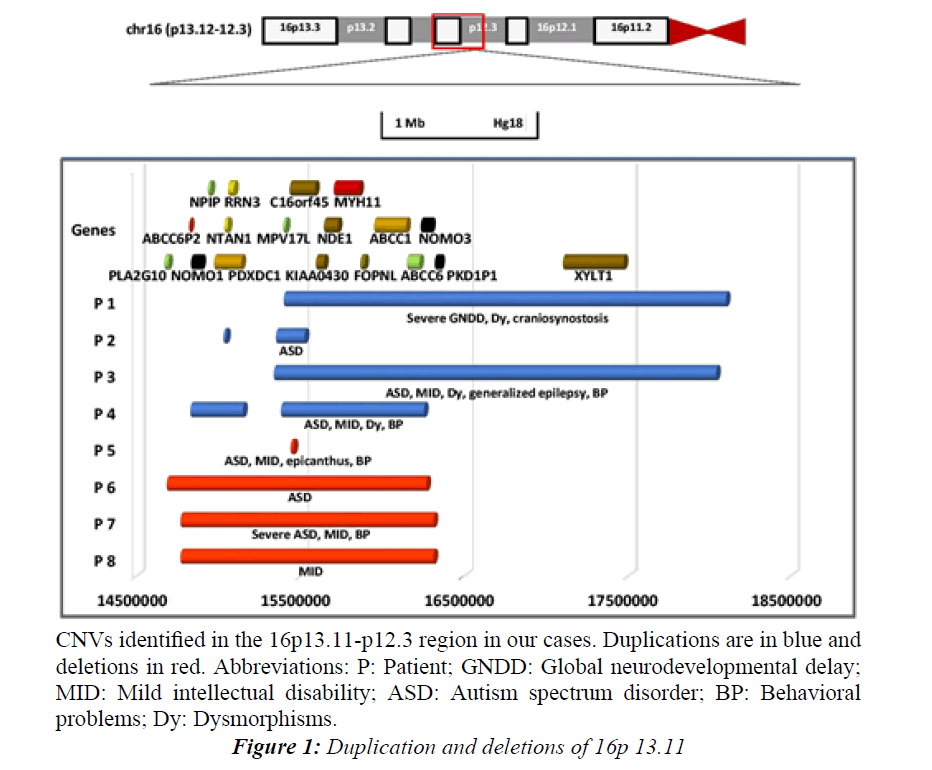
We observed that all the 16p13.11 CNVs involved the interval between C16orf45-ABCC6 (Figure 1).
Patients Carrying 16p13.11 Duplications
| Patient 1 | Patient 2 | Patient 3 | Patient 4 | |
|---|---|---|---|---|
| Gender | M | M | M | M |
| Age at array (years) | 3 | 5 | 8 | 11 |
| Clinical diagnosis Principal comorbidities |
Severe GNDD | Autism | Autism, Mild ID, generalized epilepsy |
Autism, Mild ID |
| HC (centile) | <<3 | 25 | >97 | 50 |
| Weight/Height (centile) | <3/<3 | 50/75 | 90/90 | 50/50-75 |
| Dysmorphic features Epicanthic folds/palpebral fissures/eyes Low set/posterior rotated ears Nose Mouth and palate Others |
-/up-slanting/strabismus + and dysplastic/+ - High arched palate Long philtrum, synophris |
-/-/- -/- - - - |
+/short/almond-shaped -/- Short nose, broad nasal bridge Prominent lips, macrodontia Clinodactyly, single transverse palmar crease |
-/-/- -/- - Crowding of teeth Supernumerary teeth, synophris |
| Neurodevelopmental history Regression Speech delay/Motor delay Abnormal behavior FIQ/ GDQ* level ABC (SS) |
- +/+ - - 33 |
- +/- - 104* 105 |
- Phrase speech delay/- Hyperactivity, inattention, defiance, tantrums 62* 55 |
- +/- Hyperactivity, inattention 57 65 |
| UNDA Protocol for ASD DSM-IV-TR, ADI-R, ADOS, CARS |
Negative | Positive | Positive | Positive |
| Nervous system Seizures Hypotonia/Hypertonia Structural brain anomalies |
- +/- CSA |
- -/- NA |
+ -/- - |
- -/- NA |
| Hearing/Visual deficit | -/+ | - | -/- | -/- |
| Skeletal system Hypermobility Craniosynostosis Other |
+ Trigonocephaly Long toes |
- - - |
- - Short 5th fingers |
- - - |
| Cardiovascular/other systems | Kawasaki disease/hirsutism | - | - | - |
| Family history (standardized diagnosis) |
Depression: mother | Schizophrenia: maternal grandmother | Learning difficulties: mother | Alcoholism: father; ID: first maternal cousin |
| Abbreviations: M: Masculine; GNDD: Global Neurodevelopmental Delay: ID: Intellectual Disability; HC: Head Circumference; FIQ: Full Intellectual Quotient (Wechsler [52]); GDQ: Global Developmental Quotient (Griffiths [51]); ABC: Adaptive Behavior Composite (Sparrow et al. [53]); SS: Standard Score; UNDA: Neurodevelopment and Autism Unit; DSM-IV-TR: Diagnostic and Statistical Manual of Mental Disorders, fourth edition, Text Revision (American Psychiatric Association [47]); ASD: Autism Spectrum Disorder; ADI-R: Autism Diagnostic Interview-Revised (Lord et al. [48]); ADOS: Autism Diagnostic Observation Schedule (Lord et al. [49]); CARS: Childhood Autism Rating Scale (Shopler et al. [50]); S: Social interaction; C: Communication; R: Restricted, Repetitive and Stereotyped Behaviors; DD: Developmental Delay; T: Total; CSA: Cortical and Subcortical Atrophy; NA: Not analyzed; +: Present; -: Absent; *: GDQ Level | ||||
Table 3. Clinical features of patients with duplication of 16p13.11
Patient 1: First child of non-consanguineous young parents. Apart from his mother, who had a diagnosis of depression, no other family members presented any neurodevelopmental or neuropsychiatric conditions. He was delivered by cesarean section at 38 weeks of gestational age due to acute fetal distress. The Apgar score was 5/8/10 at 1, 5 and 10 min of life, respectively. The child was reanimated with manual resuscitator. Birth weight was 2750 g (5-10th percentile), length was 46 cm (2nd percentile) and head circumference was 32.5 cm (5-10th percentile). Trigonocephaly was detected just after birth, and was submitted to surgery at seven months of age.
| Patient 1 | Patient 2 | Patient 3 | Patient 4 | |||
|---|---|---|---|---|---|---|
| Loci | 16p13.11p12.3 | 16p13.11 | 16p13.11p12.3 | 16p13.11 | ||
| Duplication length | 2.7 Mb | CNV 1: 14,8 Kb |
CNV 2: 174 Kb |
2.7 Mb | CNV 1: 0,32 Mb |
CNV 2: 0.87 Mb |
| Duplication start | 15399818 | 15032942 | 15352729 | 15339028 | 14831165 | 15387380 |
| Duplication end | 18069668 | 15047712 | 15526575 | 18035877 | 15155584 | 16256106 |
| Genes | 19 (7 MIR) MPV17L, C16orf45, KIAA0430, NDE1, MYH11, FOPNL, ABCC1, ABCC6, NOMO3, PKD1P1, LOC100288162, XYLT1 MIR-484, MIR3179-1, MIR3179-2, MIR3179-3, MIR3180- 1, MIR3180-2, MIR3180-3 |
2 PDXDC1 (last 7 exons ), NTAN1 (first 6 exons) |
3 LOC100288332, MPVL7, C16orf45 (first 2 exons) |
19 (7 MIR) MPV17L, C16orf45, KIAA0430, NDE1, MYH11, FOPNL, ABCC1, ABCC6, NOMO3, PKD1P1, LOC100288162, XYLT1 MIR 484, MIR3179-1, MIR3179-2, MIR3179-3, MIR3180- 1, MIR3180-2, MIR3180-3 |
5 NOMO1, NPIP, PDXDC1, NTAN1, RRN3 |
10 (1 MIR) MPV17L, C16orf45, KIAA0430, NDE1, MYH11, FOPNL, ABCC1, ABCC6, NOMO3, MIR484 |
| Inheritance | Maternal | Maternal | Maternal | Paternal | ||
| Other Pathogenic CNVs |
No | No | No | No | ||
| Abbreviations Mb: Megabase Pairs; CNVs: Copy Number Variations; MIR: microRNA | ||||||
Table 4: Duplication of 16p13.11: size and genes involved, based on the Human Genome HG18.
| Patient 5 | Patient 6 | Patient 7 | Patient 8 | |
|---|---|---|---|---|
| Gender | M | M | M | F |
| Age at array (years) | 4 | 14 | 14 | 12 |
| Clinical diagnosis Principal comorbidities |
Autism Mild ID |
Autism | Severe autism Mild ID |
Mild ID |
| HC (centile) | 25 | 90 | 50 | 75 |
| Weight/Height (centile) | 90/90 | 25-50/75 | 25-50/10 | 75/75 |
| Dysmorphic features | Epicanthus | - | - | - |
| Neurodevelopmental History Regression Speech delay/Motor delay Abnormal behavior FIQ/ GDQ* level ABC (SS) |
- +/- Hyperactivity, tantrums 53* 51 |
- -/- - 104 81 |
- -/- Disruptive, hyperactivity, coprolalia and OSD 52 49 |
- +/- - 58 - |
| UNDA Protocol for ASD DSM-IV-TR, ADI-R, ADOS, CARS |
Positive | Positive | Positive | Negative |
| Nervous system Seizures Hypotonia/Hypertonia Structural brain anomalies Others |
- -/- NA - |
- -/- - Clumsy |
- -/- CSA - |
- -/- NA - |
| Hearing/Visual deficit | -/- | -/- | -/+ | - |
| Skeletal system Hypermobility/Craniosynostosis Other |
-/- Pes planus, syndactyly (2nd,3th,4th toes) |
-/- - |
-/- - |
-/- - |
| Cardiovascular/other systems | - | - | - | - |
| Family history (standardized diagnosis) |
Epilepsy: mother; Depression: maternal grandmother | - | Depression: mother; Schizophrenia: paternal uncle | |
| Abbreviations: M: Masculine; F: Feminine; ID: Intellectual Disability; HC: Head Circumference; OCD: Obsessive Compulsive Disorder; FIQ: Full Intellectual Quotient (Wechsler [52]); GDQ: Global Developmental Quotient (Griffiths [51]); ABC: Adaptive Behavior Composite (Sparrow et al. [53]); SS: Standard Score; UNDA: Neurodevelopment and Autism Unit; DSM-IV-TR: Diagnostic and Statistical Manual of Mental Disorders, fourth edition, Text Revision (American Psychiatric Association [47]); ASD: Autism Spectrum Disorder; ADI-R: Autism Diagnostic Interview-Revised (Lord et al. [48]); ADOS: Autism Diagnostic Observation Schedule (Lord et al. [49]); CARS: Childhood Autism Rating Scale (Shopler et al. [50]); S: Social Interaction; C: Communication; R: Restricted, Repetitive and Stereotyped Behaviors; DD: Developmental Delay; T: Total; CSA: Cortical and Subcortical Atrophy; NA: Not analyzed; +: Present; -: Absent; *: GDQ Level | ||||
Table 5: Clinical features of patients with deletion of 16p13.11 .
Psychomotor development was delayed, and the child was referred to a neurodevelopmental evaluation. He also had recurrent ear infections, which prompted surgical treatment without residual deficit. The observation at 17 months of age revealed: axial hypotonia, no social interaction or verbal communication, poor eye contact, motor stereotypies, height, weight and head circumference under the 3rd percentile. He had strabismus, synophris, dysplastic ears, high arched palate and long philtrum (Table 3). At chronological age of four years, the VABS evaluation revealed a standard score of 33 with a homogeneous profile. He received early intervention services, physical therapy, occupational and speech therapies for language and swallowing disorders.
aCGH revealed a maternally inherited duplication of the region 16p13.11p12.3(chr16: 15399818-18069668) (Table 4 and Figure 1).
Patient 2: First born of young and healthy parents, but with a family history of schizophrenia (maternal grandmother). He was a full-term baby with an unremarkable neonatal period. He had recurring adenoiditis, which motivated surgical intervention. He developed the first words and phrases at 24th and 30th months respectively. The first symptoms were detected at 24 months of age: poor eye contact and social interaction, attachment to particular objects, pain insensitivity and problems in adapting to new environments. The physical observation was normal (Table 3). He fulfilled the UNDA protocol and was diagnosed with ASD. The GMDS revealed a global DQ of 104, a language quotient of 112 and a performance quotient of 107. The best DQ was 122 in the locomotor subscale and the worst was 88 in the eye and hand coordination subscale. His adaptive behavior composite assessed by VABS was 105 (standard score). He evolved with speech and occupational therapies and support of special education at school.
| Patient 5 | Patient 6 | Patients 7 and 8 | |
|---|---|---|---|
| Loci | 16p13.11 | 16p13.11 | 16p13.11 |
| Deletion length | 17 Kb | 1.58 Mb | 1.54 Mb |
| Deletion start | 15441629 | 14687715 | 14771033 |
| Deletion end | 15458862 | 16270740 | 16307313 |
| Genes | 1 C16orf45 |
29 (10 MIR) PLA2G10, LOC642778, LOC642799, ABCC6P2, NOMO1, NPIP, PDXDC1, NTAN1, RRN3, LOC100288332, MPV17L, C16orf45, KIAA0430, NDE1, MYH11, FOPNL, ABCC1, ABCC6, NOMO3, MIR3179-1, MIR3179-2, MIR3179-3, MIR3180-1, MIR3180-2, MIR3180-3, MIR3180-4, MIR1972-1, MIR1972-2, MIR484 |
26 (10 MIR) ABCC6P2, NOMO1, NPIP, PDXDC1, NTAN1, RRN3, LOC100288332, MPV17L, C16orf45, KIAA0430, NDE1, MYH11, FOPNL, ABCC1, ABCC6, NOMO3, MIR3179-1, MIR3179-2, MIR3179-3, MIR3180-1, MIR3180-2, MIR3180-3, MIR3180-4, MIR1972-1, MIR1972-2, MIR484 |
| Inheritance | Maternal | Maternal | Maternal |
| Other Pathogenic CNVs | No | No | No |
| Abbreviations: Kb: Kilobase Pairs; Mb: Megabase Pairs; CNVs: Copy Number Variations; MIR: microRNA | |||
Table 6. Deletion of 16p13.11: Size and genes involved, based on the Human Genome HG18.
Figure 1: Duplication and deletions of 16p 13.11.
CNVs identified in the 16p13.11-p12.3 region in our cases. Duplications are in blue and deletions in red. Abbreviations: P: Patient; GNDD: Global neurodevelopmental delay; MID: Mild intellectual disability; ASD: Autism spectrum disorder; BP: Behavioral problems; Dy: Dysmorphisms.
A maternally inherited duplication of two 16p13.11 regions (chr16:15032942-15047712 and chr16:15352729- 15526575) was identified by SNP array (Table 4 and Figure 1). The inspection with probes between the two regions confirmed the presence of two distinct CNVs.
Patient 3: First child of non-consanguineous young parents. His mother had learning difficulties. He was delivered by cesarean section at 38 weeks of gestational age, with good adaptation to extrauterine life and appropriate anthropometry.
The neurodevelopment was not of concern up to 19 months of life. At that age, he presented generalized seizures which were controlled with sodium valproate. However, he subsequently evidenced neurodevelopmental delay and macrocephaly. The electroencephalograms revealed multifocal epileptic discharges and the head Magnetic Resonance (MR) with spectroscopy was normal. The patient had rigid preferences about food and became attached to objects. Clumsy tip-toe walking, hand flapping and impaired eye contact were observed. The physical observation revealed a macrocephaly and dysmorphisms (epicanthic folds, short nose, broad nasal bridge, prominent lips, macrodontia, clinodactyly) (Table 3). At seven years old was diagnosed with ASD, according to the UNDA protocol. GMDS revealed a global DQ of 62. The language was the best area (DQ-79) and performance was the worst (DQ-49). His adaptive functioning assessed by VABS was 55 (standard score). He had sleeping and behavioral problems which motivated treatment with risperidone.
He was in school with the support of special education, speech therapy, psychomotricity, music and hydrotherapy.
aCGH detected a duplication of maternal inheritance encompassing the region 16p13.11p12.3(c hr16:15339028-18035877) (Table 4 and Figure 1).
Patient 4: First child of non-consanguineous parents, with a family history of alcoholism (father) and ID (first degree cousin, from the mother side). The neonatal period was unremarkable.
He developed with evident neurodevelopmental delay, demonstrating impairment in social interaction, poor eye contact, motor stereotypies and no verbal comprehension. His first words emerged only at the age of three and phrases at age five. Physical examination was unremarkable, with the exception of synophris and dental problems (Table 3). ASD diagnosis was confirmed. He revealed a mild intellectual disability (full intellectual quotient-IQ-57, verbal IQ-60, performance IQ-61 and standard score of the adaptive functional level-65). The hyperactive, inattentive and anxious behavior and sleep problems justified treatment with risperidone. He developed with psychological support, hydrotherapy and speech therapy. At school he had the support of special education and a functional curriculum for training the autonomy.
CNV detection by SNP array showed two consecutive duplications on 16p13.11(chr16:14831165-15155584 and chr16:15387380-16256106), which were inherited from the father (Table 4 and Figure 1). The inspection with probes between the two regions confirmed the presence of two distinct CNVs.
Patients Carrying 16p13.11 Deletions
Patient 5: First child of non-consanguineous parents, with a family history of epilepsy (mother) and depression (maternal grandmother). The delivery was by cesarean section because of acute fetal distress at 41 weeks of gestational age.
The patient developed the first words and phrases late, at age three and half and five, respectively. At age two, poor eye contact and social interaction, and motor stereotypies were observed. He presented epicanthic folds and syndactyly (Table 5). The diagnosis of ASD was made according to UNDA protocol. The GMDS revealed global DQ-53, language DQ-40 (the worst area) and performance DQ-52. The best DQ was 74 in the locomotor subscale. His adaptive behavior composite was 51 (standard score). At the behavioral level, he had tantrums and hyperactivity and sleep was problematic, motivating treatment with risperidone. He was in school with the support of special education and speech therapy.
aCGH revealed a 17 Kb maternally inherited deletion at 16p13.11(chr: 15441629-15458862) (Table 6 and Figure 1).
Patient 6: Born by normal vaginal delivery of nonconsanguineous and healthy parents. Neonatal period was marked with irritability.
Since his second year of life, difficulties in adapting to new environments, poor eye contact, deficient symbolic play and obsessions with details, especially when drawing were noticed. He presented echolalia, stereotyped verbal speech, without reciprocity, and altered prosody. He was evaluated only at eight years of age because of learning and social interaction difficulties. Physical observation was normal (Table 5). He was diagnosed with ASD without ID: full IQ-104, verbal IQ-101, performance IQ-108 and adaptive behavior composite of 81 (standard score). The academic progress was regular at school, with a music program.
A maternally inherited deletion 16p13.11 (chr16:14687715- 16270740) (Table 6 and Figure 1) was detected by SNP array.
Patient 7: First of two children from young non consanguineous parents. The neonatal period was unremarkable. He had a diagnosis of myopia and a positive family history of depression (mother), schizophrenia (paternal uncle) and ID (sister).
The first neurodevelopmental difficulties started at 24 months of age when he evidenced hyperactivity, motor stereotypies and repetitive speech. Physical observation was normal (Table 5). He fulfilled the UNDA protocol and was diagnosed with severe ASD and mild ID (full IQ- 52, verbal IQ-57, performance IQ-51, adaptive behavior composite-49). He developed with obsessive behavior, defiance, verbal and physical aggressiveness and coprolalia. At 15 years old, he developed an obsessive-compulsive disorder that motivated a referral to psychiatric evaluation and multiple medications (risperidone, olanzapine and fluoxetine).
CNV detection by SNP array revealed a maternally inherited deletion encompassing the region 16p13.11(c hr16:14771033-16307313) (Table 6 and Figure 1).
Patient 8: This girl is the sister of patient 7. The delivery was by cesarean section because of acute fetal distress at 40 weeks of gestational age. The Apgar score was 4/8/10 at 1, 5 and 10 min of life, respectively, and was reanimated with manual resuscitator.
She was evaluated because ID was suspected based on learning difficulties, retrospective history of language delay that motivated speech therapy and familial history. She had no stigmas of ASD. The physical observation was normal (Table 5). The evaluation was compatible with mild ID (full IQ-58, verbal IQ-53 and performance IQ-72).
She was submitted to molecular analysis due to ID and a positive family history of ASD (brother) and the SNP microarray analysis revealed a maternally inherited deletion of the following genomic region: 16p13.11(c hr16:14771033-16307313) (Table 6 and Figure 1).
Discussion
We describe eight pediatric cases with 16p13.11 microdeletions/microduplications and neurodevelopmental disorders, six with ASD and two with GNDD/ID. All the CNVs were inherited.
The sizes of the rearrangements were variable, ranging from 17 Kb to 2.7 Mb. The majority of the CNVs encompassed the ~15400000-16200000 region of 16p, which include the C16orf45, KIAA0430, NDE1, MYH11, FOPNL, ABCC1, ABCC6 genes (Figure 1).
The clinical manifestations associated with 16p13.11 duplications are in agreement with the neurodevelopmental phenotype previously described, namely the association with ASD, ID, dysmorphisms and behavioral problems [26-28,31]. The dysmorphisms were only observed in patients with duplication. The behavioral problems were observed in the same proportion of duplications and deletions (2 cases in each group).
Generalized epilepsy is more frequent in cases with deletion of the 16p13.11 region, that is considered the most prevalent single genetic risk factor for overall seizure susceptibility identified to date, but there are cases described in association with the duplication, like our case [31,38-40]. The presence of cardiac involvement and skeletal manifestations were observed in other series but we described only one patient with craniosynostosis [26,37]. In contrast to the literature, we did not have patients with 16p13.11 deletions and microcephaly.
In our study, the variable expressivity is evident and, interestingly, the majority of CNVs was transmitted from parents with neuropsychiatric or cognitive phenotypes including depression, learning problems or alcoholism. The genetic or epigenetic mechanism for the variable expressivity in the mutation-carrying parent is not known [8]. Possibly, the 16p13.11 CNVs explain only part of the phenotypic variability. Additional factors, either genetic or epigenetic, like methylation patterns, or environmental factors, are likely to ultimately influence the phenotypic expression of variability in these loci [33,41,55]. Additional genetic factors, such as other pathogenic CNVs, may be identified via CMA, but in many cases they are not found, like in our patients and in other series [26,56].
In our study, the majority of the pediatric patients were males. In fact, recently, Tropeano et al. [29] and Pinto et al. [19], reported evidence for a male-biased effect of 16p13.11 CNVs and the same pattern of results was also observed in the Decipher sample [57]. Patients 1-3 and 5-7 are all males with maternally inherited events. This is a fairly dramatic pattern in seven families. Possibilily, female's are more "protected" from susceptibility of this neurobehavioral phenotype and as a result the manifestations are often seen in their male offspring. This pattern is supported by other publications, but the mechanism of the female protective effect remains unknown [58,59].
The sex-limited effect on the penetrance of the pathological phenotypes, could result from imprinting, endocrine, impaired interhemispheric connectivity, in males and requirement of higher genetic load to express ASD, in females [19,29]. Despite the cause, it is obvious a categorical protection from expression in females (resilience) like in other types of neuropathology [29].
However, it will be necessary to perform extended family studies to confirm these data. Ullman et al. has previously hypothesized that duplications that are paternally transmitted are benign while the maternal transmission leads to clinical manifestations [28]. However, there are no known or predicted imprinted genes in this region and here we describe clinical manifestations in patients with duplications inherited from both parents, as did Nagamani et al. [26].
Our cases highlighted the concept of the pleiotropy: CNVs predisposes to a myriad of neurodevelopmental signs and symptoms, including epilepsy, as well as to physical abnormalities [13,34]. Findings of several studies strengthen the hypothesis that there are shared genetic risk factors between schizophrenia, psychosis, epilepsy, ID, autism and OCD [27,28,34,35,38-40,45,46,60]. These data imply the existence of shared biologic pathways among multiple neurodevelopmental conditions [34].
An analysis of the reported cases with 16p13.11 CNVs, defines a common chromosomal region encompassing several genes: C16orf45, KIAA0430, NDE1, MYH11, FOPNL, ABCC1 and ABCC6 genes (Figure 1).
Two of these genes, NDE1 (nudE nuclear distribution gene E homolog 1) and NTAN1 (N-terminal asparagine amidase), have been implicated in the neurocognitive phenotype [26,39,40,61,62]. Although the loss of copy number in these genes has resulted in neurological manifestations in animal models, the phenotypic consequences of gain of copy number is still unclear. Thus, the role of duplication of these genes in behavioral and cognitive impairments is not well established. However, it is not unusual for reciprocal deletions and duplications to present with overlapping phenotypes [26]. Conversely, in our study, not all the CNVs had involvement of NTAN1 gene and of the NDE1 genes, highlighting the importance of other genes in this region. Bioinformatics analysis of the gene content of the 16p13.11 region in one study, found evidence that several of the genes in the region, such as ABCC1, NOMO1, NTAN1 and PDXDC1 are expressed during brain development as is NDE1 [44].
Recently, Fujitani and colleagues (2016) identified miR- 484 gene as the responsible gene for the behavioral hyperactivity phenotype in 16p13.11 microduplication [63]. Moreover, they found that miR-484 controls cortical neurogenesis by inhibiting protocadherin-19 (PCDH19) translation and mRNA stabilization [63]. In our cases, the hyperactive phenotype was observed both in cases with duplication and with deletion. The miR-484 gene was involved in all of them except for one case (Patient 5).
Zhang et al. (2014), reported a role of another susceptible gene 2900011O08Rik, which is also located on the 16p13.11 locus and was named gene MINP (Migration Inhibitory Protein) according to its distinct role in the developing cortex [64]. This gene, that is the mouse homolog of C16orf45 gene, is specifically expressed in the nervous system and is highly expressed in postmitotic neurons of the cerebral cortex. Moreover, in vivo downregulation of this gene specifically accelerated radial migration without changing the neuronal morphology and cortical lamination [64]. C16orf45 gene- chromosome 16 open reading frame 45 (a protein-coding gene) - was the only gene involved in all of our cases. Patient 5, who had ASD, mild ID and behavioral problems, carried a 17 Kb deletion of maternal origin spanning only the C16orf45 gene and, interestingly, his mother had epilepsy. In the Database of Genomic Variants (DGV) there are only three reports of deletions in this region in normal individuals [54]. Hence, this small deletion, might display reduced penetrance. Further, it was one of the only five genes involved in one case with reciprocal duplication (Patient 2), defining a minimum critical region.
Little is known about the role of the C16orf45 in the phenotypic spectrum of 16p13.11 rearrangements. In one hand, there is evidence that there is interplay between C16orf45, a well conserved gene in mammalians, highly expressed in the brain, and the other genes in region, as observed by accessing to STRING10 database [57,65,66]. In the other hand, there were some cases reported in the Decipher Database involving this gene, but larger than this CNV [57,67].
In fact, the overwhelming majority of CNVs in this region arise from non-allelic homologous recombination between adjacent segmental duplications [26,27,29-31]. In contrast, CNVs of patients 2 and 5 are almost entirely within the repetitive genomic region and need differentiated analysis. Considering that CNV of patient 2 involve only C16orf45, it could be seen as a nested duplication, and be useful as a tool to identify which of the genes (or set of genes) would be responsible for the phenotype. The breakpoints of this CNV lie within the gene and could be acting as a duplication (and hence possibly being expressed at higher levels than controls) or as a loss-of-function allele, as an intragenic inverted duplication could disrupt the reading frame and effectively act as a deletion.
This might indicate that C16orf45 has an important role in neurodevelopment and in the phenotype observed in these patients. The study findings of Zhang et al. laid the foundation for exploring the function of the novel gene MINP ( C16orf45) [64]. Further analysis for uncovering other distinctive roles of MINP in the nervous system is currently in progress. Because MINP is a risk gene located at 16p13.11, transgenic animals carrying MINP mutations are expected to serve as new models for neuropsychiatric diseases associated with 16p13.11 CNVs [64].
However, several possibilities might not involve c16orf45, like CNVs disrupting expression of more well established genes in the region such as NTAN1 which could be a consequence of the patients´ CNVs, including the CNV of the patient 5. Other hypothesis is that each patient has a different etiology leading to some common patterns of neurobehavioral and neurocognitive phenotypes. Other possibility is that given