- Biomedical Research (2011) Volume 22, Issue 1
Comparative Analysis of Effects of Urea and Trypsin on Human Chromosome Banding
Yogesh AS*, M Joshi, RR Marathe and J DeshpandeDepartment of Anatomy, J N. Medical College, Sawangi (Meghe), Wardha, MS, India
- *Corresponding Author:
- Yogesh A S
Cytogenetic Laboratory
Department of Anatomy
J N Medical College
Wardha 442004, M.S
India
Accepted date: September 23 2010
Abstract
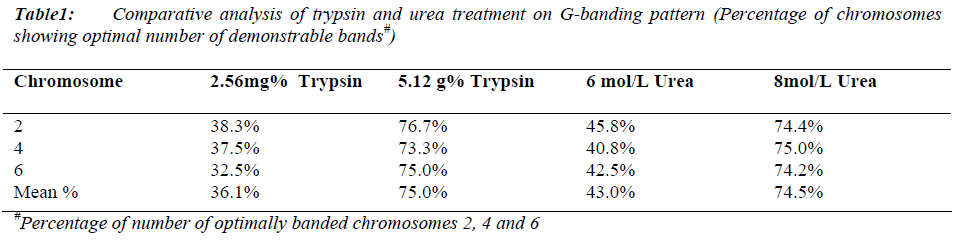
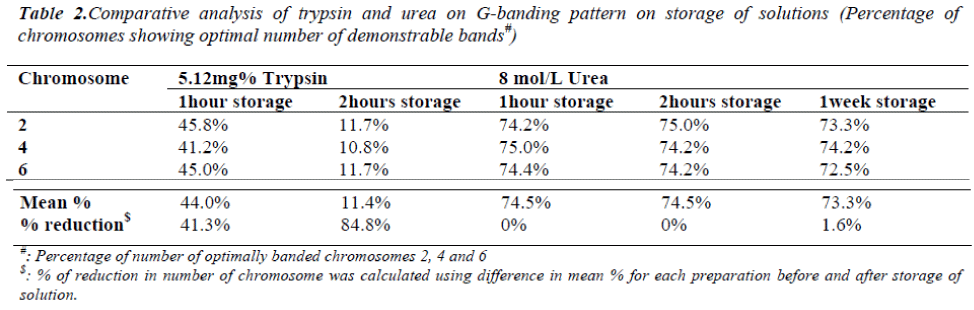
Obtaining good banding pattern of human chromosomes is always a critical step in cytogenetic analysis. Effects of various concentrations of urea and trypsin on human chromosome banding were studied. Maximum percentage of chromosomes showing optimal number of demonstrable bands was obtained with 5.12mg% freshly prepared trypsin solution (pH 6.8) in 16 to 18 seconds for aged slides as well as with 8 mol/L fresh urea solution in 30 to 90 seconds for freshly prepared slides. There was no resolution difference at these concentrations. The percentage of chromosomes showing optimal number of demonstrable bands were decreased by 41.3% and 84.8% on one hour and two hours storage of 5.12mg% trypsin solution at 370C respectively with severe effect on band resolution. While no difference was found on one hour and two hours storage of 8 mol/L urea solution at 370C and even on storage for a week at 40C there is only 1.6% decrease without affecting band resolution. Thus urea method offers an advantage in having better stability of solution and non requirement of slide ageing while longer urea treatment time is a disadvantage.
Keywords
G-banding, urea, trypsin
Introduction
After treatment of proteolytic agent, chromosomes reveal characteristic patterns of horizontal dark and light bands like bar codes on staining. The banding patterns lend each chromosome a distinctive appearance so the chromosomes can be identified and distinguished without ambiguity. Banding also permits the recognition of chro-mosome deletions (lost segments), chromosome duplica-tions (surplus segments) and other types of structural rearrangements of chromosomes. The existence of speci-fic banding patterns which enable individual human chromosomes to be identified was first demonstrated by Caspersson et al in 1968 using quinacrine fluorescence [1]. But the minimum constrain of this method was use of fluorescence microscopy. In Giemsa banding method (G-banding), chromosome slides are treated with proteolytic agents followed by staining with Romanowaski dyes. Seabright has described the trypsin for G-banding produc-tion [2]. Many researchers studied the use of other proteolytic agents such as papin [3], urea [2].
Although the exact molecular mechanisms involved in the production of G-bands still remain unknown, it is possible to summarize that the G-bands are the result of dissociation of protein in the chromosome structure. The
darkly stained regions represent an undenaturated DNA component of the chromosome. Heterochromatin at secondary constrictions also produces dark bands. Site for the proteolytic agent during G-banding are the Histone proteins. The probable reason behind the positive G-banding is the compact structure of chromosome at G-C rich regions where histone proteins are not exposable to proteolytic agent [4]. Trypsin is most commonly used agent for G-banding at the concentration 5.12 mg% in phosphate buffer pH 6.8 to 7.6 [5]. Urea shows proteolytic activity at 8 mol/L concentration [6]. Till date difficulty in obtaining good banding results is continued. There are several factors affecting the appearance of chromosome banding such as proteolytic agent, drying of the slide, ageing of the slide, colchicine exposure time, agents inhibiting chromosome contraction such as ethidium bromide, actinimycin D, fixative, hypotonic saline treatment, duration of fixation, staining solutions and its pH [5]. Due to all these aspect, to obtain good quality banding pattern is tedious job for laboratory personnel. Keeping all other conditions similar, in the present study we have done the comparative analysis on the effects of the various concentrations of urea and trypsin and effect of storage of their solutions on human chromosome banding.
Material and Methods
Blood sample (3 ml) from 10 healthy individuals was collected in preheparinized syringes. Each sample of peripheral blood was processed for cytogenetic analysis as described earlier [7]. In brief, blood sample was cultured in RPMI1640medium (Himedia) supplemented with 10% fetal bovine serum (Himedia) and 10 μg Phytohemagglutinin /ml (Genei) and incubated at 37°C for 72 hrs. One hr and 20 min prior to harvesting; 100 μL Colchicine (Himedia) (0.25 μL/ml) was added to each culture tube. Cells were centrifuged at 1000rpm for 10min. Cells were suspended in 10 ml of prewarmed (37°C) 0.075mol/L KCl (hypotonic solution) and incubated at 37°C for 10min. Cells were resuspended in 5 ml of chilled fixative (methanol: glacial acetic acid:: 1: 3). The fixative was removed by centrifugation and the procedure was repeated thrice. To prepare slides 2-3drops of fixed cell suspension were dropped on clean slides and air dried.
Trypsin method: Slides aged at 950C for 25 min (thermal ageing method) were treated with 2.56 mg% and 5.12 mg% freshly prepared, prewarmed trypsin (1:250, Himedia) solutions (6.8 pH) for 12 -20 seconds. For trypsin inactivation, the slides dipped into chilled normal saline for 10 seconds. Then the slides were stained with Leishman stain and observed under microscope. After
storage of 5.12 mg% trypsin solution for 1hour and 2 hours, same procedure was followed for banding.
Urea method: Freshly prepared slides were treated with 6M and 8M urea solutions for 30-90 seconds and washed thoroughly with distilled water. Then the slides were stained in Leishman stain. After storage of 8M urea solution for 1hour and 2 hours at 37°C and for 1week at 4°C, same procedure was followed.
Slides were screened using Olympus CX31 (Japan) microscope and microphotographed. Data was analyzed using SPSS version 14.0 software. Mitotic index (Mitotic Index = Number of cells in mitosis per 1000 observed cells) was determined to rule out the sampling error. The results for each preparative method were assessed by comparing the banding patterns of G-banded chromoso-mes at 550 band level suggested by ISCN-2005 [8]. In each preparation, 20 metaphases with good chromosome separation were studied. Chromosome number 2, 4 and 6 are the better for assessing the banding effect [9]. Number of optimally banded chromosomes 2, 4 and 6 were noted from 20 metaphases from each preparation.
Results
Mean mitotic index was 112/1000 cells (maximum: 116/1000 and minimum: 108/1000 cells) with no significant difference in all samples (p > 0.001) (Data not shown). The maximum percentage of chromosomes 2, 4 and 6 showing optimal number of demonstrable bands were obtained with 5.12mg% fresh trypsin solution in 16 to 18 seconds for slides aged as well as with 8mol/L fresh urea solution in 30 to 90 seconds for freshly prepared slides (Table 1). There was no resolution deference in both conditions.
On one hour and two hours storage of 5.12mg% trypsin solution at 37°C, the percentage of chromosomes showing optimal number of demonstrable bands were decreased by 41.3% and 84.8% respectively (Table 2). This decrease was found to be significant (p<0.001). The band resolu-tion of most of the chromosome bands was severely affected on trypsin solution storage for two hours. On one hour and two hours storage of 8mol/L urea solution at 37°C, the percentage of chromosomes showing optimal number of demonstrable bands was found same. Even on storage for a week at 4°C, decrease was only 1.6% (Table 2) which was non-significant (p>0.001). The band reso-lution was not affected on urea solution storage even for a week.
Discussion
Cytogenetic analysis in many clinical cases such as Down’s syndrome (trisomy 21), Klinefelter's syndrome (47,XXY), Turner’s syndrome (45,X), Ph chromosome [(t(9;22)(q34;q11)] routinely depends upon identification of G-banded chromosomes. During G-banding, the site of activity of proteolytic agent is the histone protein. These proteins complex with DNA in the nucleosome structure, in which two molecules of each of the histones H1A, H2B, H3, and H4 form an octamer with two turns of double helix DNA wound around this histone octamer [10]. The nucleosomes are connected by DNA linkers and are bound by histone H1 in a 30-nm fiber, which is observed both in interphase chromatin and metaphase chromosomes [11]. Positive G-bands are located in short and long arm of all chromosomes but they are located invariably at heterochromatin [8]. Positive G-banded regions contain Guanine-Cytosine (dG-dC)-enriched DNA sequences and replicate in late S-phase [4]. According to Comings, constitutive heterochromatin may be dGuanine-dCytosine rich [4].
In the present study, both the methods have shown their own advantages and disadvantages. Trypsin is a serine protease that cleaves peptide chains at the carboxyl side of the amino acids lysine or arginine. Trypsin has an optimal operating pH of about 8 and optimal operating temperature of about 37°C. As trypsin exhibits autoproteolytic activity, every time fresh solution has to be prepared [12]. Urea is a byproduct of nitrogen metabolism; having proteolytic activity. As urea does not show autolysis activity, its solution may be useful even after 1 week of preparation. Trypsin is more active even at lower concentrations and unfavorable pH as compared to urea.
It is because trypsin acts enzymatically and urea is protein denaturant [6,12]. To reduce the vigorous activity of the trypsin, in the present study, for the solution of trypsin pH 6.8 is used instead of its optimal pH. As a fresh learner takes time to achieve the proficiency of G-banding technique, trypsin solution may become useless. Hence, in such teaching laboratories, the urea method may be useful. While in clinical laboratories, trypsin method may be more useful due to less time consumption during each slide preparation.
Acknowledgement
This study was supported by Datta Meghe Institute of Medical Sciences University, Nagpur, India. Thanks are due to Honorable Dr. Vedprakash Mishra, Vice-chancellor and Dr. SS Patel, Chief Coordinator, DMIMS University for keen interest and encouragement; Dr. VK Deshpande, Dean, JN Medical College, Wardha, for cooperation.
References
- Caspersson T, Farber S, Foley GE, Kudynowski J, Modest EJ, Simonsson E, Wagh U, Zech L. Chemical differentiation along metaphase chromosomes. Experimental Cell Research 1968; 49, 219-222.
- Seabright M. A rapid banding technique for human chromosomes (Letter). Lancet 1971; 2: 971-972.
- Dutrillaux B, de Grouchy J, Finaz C, Lejeune J. Mise en evidence de la structure fine des chromosomes humains par digestion enzymatique (pronase enparticulier). Comptes rendus hebdoma daires des Seances de l'Academie des Sciences, D 1971; 273: 587-588.
- Comings DE. Mechanisms of chromosome banding and implications for chromosome structure. Ann Rev Genet 1978; 12: 25.
- Schreck RR, Distèche CM. Chromosome banding techniques: In Current Protocols in Human Genetics. Dracopoli NC, Haines JL, Korf BR, et al, eds. John Wiley Current Protocols DOI: 10.1002/0471142905.hg0402s00 last update Jan 14, 2010.
- Rossky PJ. Protein denaturation by urea: Slash and bond. Proc Natl Acad Sci USA. 2008; 105: 16825-16826.
- Yogesh AS, RR Fulzele. Cytogenetic study on genotoxicity of antitumor-antibiotic Mitomycin C. Biomed Res 2009; 20(1): 40-44.
- Shaffer LG, Tommerup N eds. ISCN-2005: An International system for human cytogenetic nomenclature, S.Karger, Basel, published in collaboration with ‘cytogenetic and genomic research’ 2008.
- Barch MJ, Knutsen T, Spurbeck JL, eds. The AGT cytogenetic manual. 3rd Ed. Lippincott Raven New York, 1995; pp-278.
- Heinrichs A. Chromatin: A positive twist. Nat Rev Mol Cell Biol 2009; 10: 582. doi:10.1038/nrm2750
- Rando OJ, Chang HY. Genome-Wide Views of Chromatin Structure. Anal Rev Biochem 2009; 78:245-271. doi:10.1146/annurev.biochem.78.071107.134639
- Lee WS, Park CH, Byun SM. Streptomyces griseus Trypsin is Stabilized against Autolysis by the Cooperation of a Salt Bridge and Cation-Π Interaction. J Biochem 2004; 135(1): 93-99.