- Biomedical Research (2014) Volume 25, Issue 1
Chemical fingerprinting of Saudi Arabian snake venoms using gel filtrationchromatography.
Abdulrahman Al Asmari1*, Haseeb Ahmad Khan2 and Rajamohammed Abbas Manthiri11Research Center, Prince Sultan Military Medical City, Riyadh, Saudi Arabia
2Department of Biochemistry, College of Science, King Saud University, Riyadh, Saudi Arabia
- *Corresponding Author:
- Abdulrahman Al-Asmari
Director of Research Center
Prince Sultan Military Medical City
P.O. Box 7897, Riyadh 11159, Saudi Arabia
Accepted date: November 07 2013
Abstract
Snake bite is crucial medical emergency while the success of immediate care and anti-venom therapy mainly depend on the correct identification of culprit snake. We present a simple method for identification of six snake species including Echiscoloratus, Echispyramidum, Cerastescerastesgaspreti, Bitisarieatans, Najahaje Arabica, andWalterinnesiaaegyptia, using the gel filtration chromatographic profiles of their venoms. The chromatograms of venoms from different snake species showed peculiar patterns based on the number and location of peaks.Our findings suggest that chromatographic profiles of snake venoms provide a simple and reproducible chemical fingerprinting method for quick identification of snake species.
Keywords
Snake venom, Chromatographic profiles, Elapidae, Viperidae, Identification
Introduction
The venoms from Elapidae and Viperidae snakes are complex mixturescontaining different components such asmetalloproteinases, proteolytic enzymes, phospholipase, serine proteinase, presynaptic and postsynaptic neurotoxins, potassium channel-binding neurotoxins, cytotoxins, cardiotoxins and platelet aggregation inhibitors [1-3].Newton et al [4] have suggested that analysis of venom components can produce a unique fingerprint to be used as a valuable reference tool in taxonomic analysis asa complementary method to morphology and behavioral characterization for species identification and classification. Compositionaldifferences between snake venoms can be employed as a taxonomy signature forunambiguous species identification independently of geographic origin andmorphological characteristics [5]. Recently, Calvete [6] have pointed out that use of proteomics approaches in identification of evolutionary and immunoreactivitytrends among homologous and heterologous venoms may aid in the replacement of thetraditional geographic- and phylogenetic-driven hypotheses for antivenom production strategies.
In continuation to our previous work on chemical fingerprinting of scorpion venoms [7], we performed the gel filtration chromatography of snake venoms from four species from the family Viperidae (Echiscoloratus, Echispyramidum, Cerastescerastesgaspreti and Bitisarieatans) and two species from the family Elapidae (Najahaje Arabica and Walterinnesiaaegyptia) and compared the chromatographic profiles for their application in species identification.
Materials and Methods
We collected six species of snakes (Fig. 1) including Echiscoloratus, Echispyramidum, Cerastescerastesgaspreti, Bitisarieatans, Najahaje Arabica and Walterinnesiaaegyptiafrom the Riyadh region of Saudi Arabia. The snakes were kept in plastic boxes and fed onmice and water adlibitum. The crude venom was diluted with distilled water, properly mixed, and centrifuged at 10,000 rpm at 4°C for 20 min to separate the mucus. The clear supernatant was filtered through 0.20 μm filter before chromatography.
Gelfiltration chromatography was used for venom fractionation on Superdex200 PC 3.2/30 column. Venom solution was diluted in 0.05 M sodium phosphate buffer containing 0.15 M NaCl (pH 7.0). An aliquot (25μl) of venom solution was loaded in a previously equilibrated column with the same buffer. The sample was injected using an Auto injector. The flow rate was adjusted at 0.4 mL/min and the UV range was 0.02 AUFS (absorbance unit full scale). Column operational pressure was 1.5 MPa. The elution profile was monitored at 280 nm by a UV spectrophotometer (AKTA Micro System). All the samples were run in triplicate to confirm the reproducibility of their chromatographic patterns.
Results and Discussion
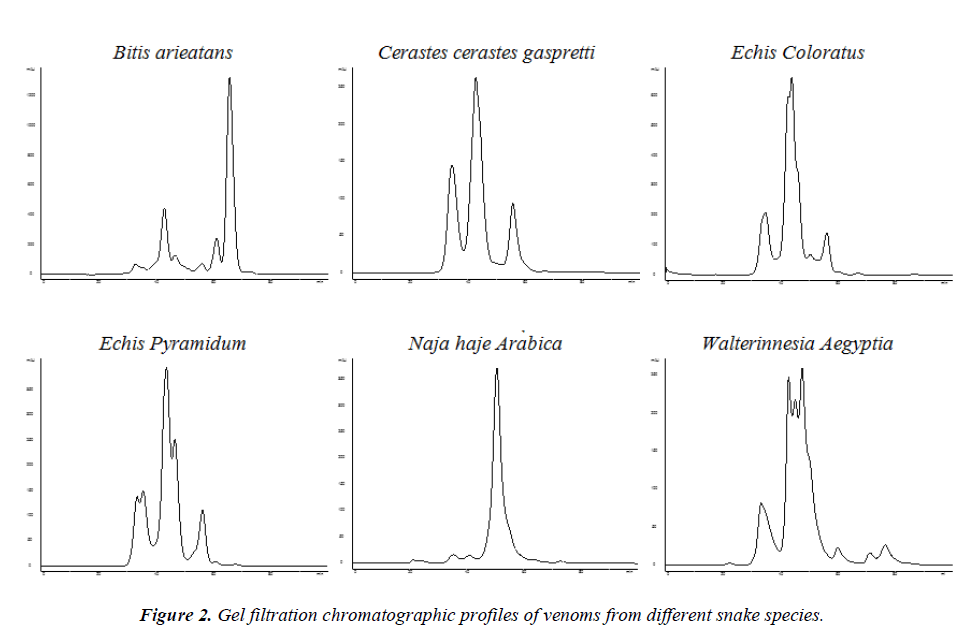
All the snake species showed peculiar chromatographic profiles of their venoms depending on the location and height of the peaks (Fig 2). The minimum numbers of peaks were observed with the venom of Cerastescerastesgaspreti ( 3 peaks) and the maximum number of peaks with the venom of Walterinnesiaaegyptia (7 peaks). Both the members of the genus Echis (Echiscoloratus and Echispyramidum)showed 5 peaks each (3 common peaks) whereas the venoms of Najahaje Arabica and Bitisarieatans resulted in 4 and 6 peaks respectively (Fig. 2).
The venom profile-based chemical fingerprinting clearly differentiated the two species of the family Elapidae (Naja Arabica and Walterinnesia Aegyptia) from the members of the family Viperidae. The venom profile of Echiscoloratus was more closely related to the venom profile of Cerastesgaspreti instead of Echispyramidum (Fig. 2). Aird [8] compared the gel filtration profiles of crude snake venoms from 38 Crotalusviridis,representing the subspecies concolor, viridis and lutosus and readily distinguished these taxa. John and Kaiser [9] conducted a comparative study of the venoms from Notechisscutatusscutatus, Notechisaterserventyi, Notechisaterhumphreysi and Notechisateraterusing gel filtration resulting in slightly different elution profiles on a Superose-12 gel filtration column.The protein profile of venoms of Elapidae was identified using electrofocusing technique; the two species could easily be differentiated whereas the differences between the two sub-species were more difficult to evidence [10]. The elution profiles of the venoms of seven Bothrops species fractionated on a Mono-Q FPLC column resulted in reproducible chromatogramshowever there was a considerable overlap of activeproteins in different species venoms [11].
In conclusion, eachsnake’s venom has a unique chromatographic profile that can be used as a fingerprint to differentiate one speciesfrom the other.Our findings suggest that gel filtration chromatography of snake venom is a simple and reproducible method for identification of snake species.
Acknowledgments
The authors extend their appreciation to the Deanship of Scientific Research at King Saud University, Riyadh, Saudi Arabia for funding the work through the research group project No. RGP-VPP-009.
References
- Tu AT. Overview of snake venom chemistry. AdvExp Med Biol 1988; 391: 37-62.
- Meier J, Stocker K. Effects of snake venoms on hemostasis. Crit Rev Toxicol 1991; 21:171-182.
- Fry BG. From genome to ‘venome’: molecular origin and evolution of the snake venom proteome inferred from phylogenetic analysis of toxin sequences and related body proteins. Genome Res 2005; 15: 403-420.
- Newton KA, Clench MR, Deshmukh R, Jayaseelan K, Strong PN. Mass fingerprinting of toxic fractions from the venom of the Indian red scorpion, Mesobuthustamulus: biotope-specific variation in the expression of venom peptides. Rapid Commun Mass Spectrom 2007; 21: 3467-3476.
- Tashima AK, Sanz L, Camargo AC, Serrano SM, Calvete JJ. Snake venomics of the Brazilian pitvipers-Bothropscotiara and Bothropsfonsecai. Identification of taxonomy markers. J Proteomics. 2008; 71: 473-485.
- Calvete JJ. Snake venomics: From the inventory of toxins to biology. Toxicon. 2013 Apr 9. pii: S0041-0101(13)00115-3.
- AlAsmari A, Khan HA, Manthiri RA. Rapid profiling of crude scorpion venom using liquid chromatography and its relevance to species identification. ActaChromatogr 2012; 24: 501-509.
- Aird SD. A quantitative assessment of variation in venom constituents within and between three nominal rattlesnake subspecies. Toxicon 1985; 23: 1000-1004.
- John TR, Kaiser II. Comparison of venom constituents from four tiger snake (Notechis) subspecies. Toxicon 1990; 28: 1117-1122.
- Pichon-Prum N, Debourcieu L, Chaperon N. Comparison of two Elapidae venoms: Najanaja and Najanigricollis. Ann Pharm Fr. 1990; 48: 87-93.
- Leite LC, Furtado MF, Correa TC, Raw I. Characterization of the snake venoms from seven Brazilian species of Bothrops by FPLC anion-exchange chromatography. CompBiochemPhysiol B 1992; 102: 515-520.