Brief Report - Archives of General Internal Medicine (2023) Volume 7, Issue 4
Brief review of miRNAs as new and natural biomarkers in the treatment of type 2 diabetes mellitus
Ali Salehi*Department of Cellular and Molecular Biology, Islamic Azad University, Esfahan, Iran
- *Corresponding Author:
- Ali Salehi
Department of Cellular and Molecular Biology
Islamic Azad University, Esfahan, Iran
E-mail: salehiali618@gmail.com
Received: 22-Jul-2023, Manuscript No. AAAGIM-23-106586; Editor assigned: 24-Jul-2023, PreQC No. AAAGIM-23-106586(PQ); Reviewed: 07-Aug-2023, QC No. AAAGIM-23-106586; Revised: 10-Aug-2023, Manuscript No. AAAGIM-23-106586(R); Published: 17-Aug-2023, DOI:10.35841/aaagim-7.4.181
Citation: Salehi A. Brief review of miRNAs as new and natural biomarkers in the treatment of type-2 diabetes mellitus. Arch Gen Intern Med. 2023;7(4):181
Abstract
The prevalence of type-2 diabetes as a progressive metabolic disorder is rapidly increasing worldwide. Diagnosing this disease in the early stages (pre-diabetes) plays an important role in reducing deaths related to this disease. Studies have shown that micro RNAs play an important role in the pathogenesis of type-2 diabetes. Due to the stability of these regulatory molecules in body fluids and the availability of sensitive technology for their measurement, they can be used as genetic biomarkers in the early diagnosis of type-2 diabetes. We made a new diagnosis of disease
Keywords
Diabetes mellitus, miRNA, Biomarker.
Introduction
Diabetes mellitus is a group of complex and diverse metabolic disorders related to age [1,2]. This disease is characterized by insufficient secretion of insulin from the beta cells of the pancreas, which prevents normal blood glucose homeostasis. Type-1 diabetes is caused by a lack of production of the insulin hormone from the beta cells of the pancreas, while type-2 diabetes is caused by an ineffective insulin response. Diabetes type 1 and diabetes type 2 are different diseases in terms of pathogenesis and treatment [3]. Examples of secondary complications of diabetes include neuropathy, retinopathy, nephropathy, atherosclerotic disease, ischemia of the heart, cerebral vessels, etc. Diabetes and its secondary complications are the main cause of disability, reduced quality of life and early death [4,5]. The current treatments available for this disease are not completely effective, that is why there is a need to better understand this disease and find new treatment methods and new markers to identify this disease, knowing the molecular mechanisms underlying diabetes can be used as new markers and the new therapeutic goals of this disease [6].
miRNAs are small non-coding single-stranded molecules with a length of about 22 nucleotides [7]. These molecules regulate gene expression after transcription by binding to the 3' untranslated region of messenger RNA, thus preventing translation or mRNA degradation. miRNAs play a key role in regulating cellular functions such as apoptosis, differentiation, proliferation, carcinogenesis, glucose homeostasis, inflammation, and other biological processes [8-11]. Since miRNAs play a key role in physiological processes and can modulate pathophysiological disease states, there is an urgent need to identify miRNAs and their targets in relation to diabetes complications, as it provides a new opportunity to identify new biomarkers and therapeutic targets. He does. While several studies have shown that miRNAs may play a role in diabetes itself and vascular complications not related to diabetes [12-16]. In recent years, studies have shown that circulating miRNAs are related to various diseases, including diabetes, for this reason, the knowledge of circulating miRNAs may reflect the dynamic changes of cells in response to the disease, and because of this, their potential as markers. Biological methods for diagnosing and predicting diabetes have been increasingly considered [17]. For this reason, in this review, we will examine the role of miRNAs in diabetes and examine them as biomarkers for this disease.
Check method
To carry out this study, 63 sources of original articles, metaanalysis and review articles related to diabetes mellitus and its biomarkers, including miRNA, were reviewed. It was searched in the databases of Google Scholar, Elsevier, PubMed, and...
Examining the function of miRNAs in diabetes disease
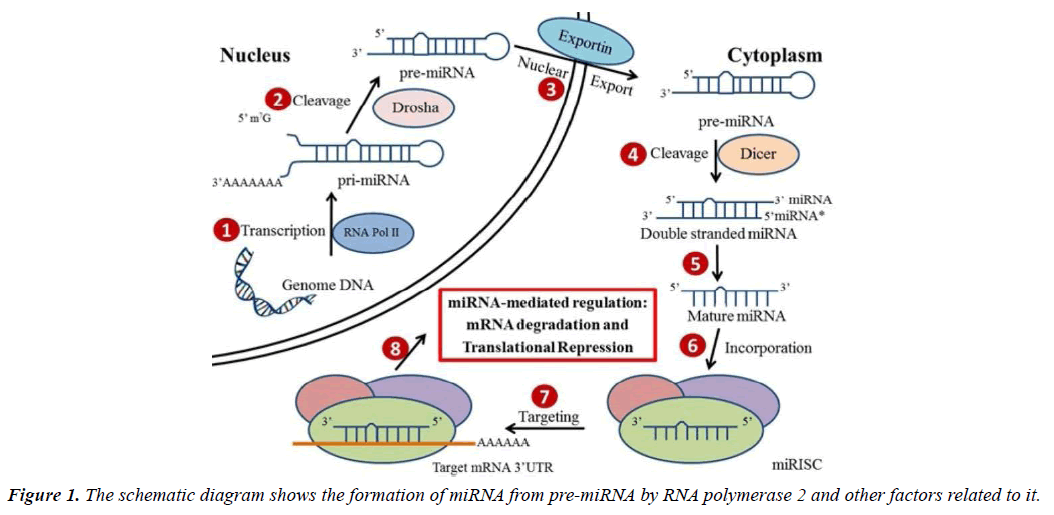
miRNA biogenesis: microRNAs are short non-coding RNAs, which are produced endogenously and are about 20 to 22 nucleotides in length, which play an important role in modulating mammalian genes and regulate several cellular functions [18-23]. Found in plants, animals, viruses, human tissues and blood, more than 2000 different miRNAs have been found in the human genome. Even though most of the mature miRNAs are approximately 22 nucleotides in length, they are first transcribed into long precursor molecules called primary miRNAs (pri-miRNAs) by RNA polymerase II enzyme. Then with the help of nuclear microprocessor complex consisting of RNase III enzyme Drosha, the pri-miRNAs are further transformed into smaller hairpin-shaped precursor miRNAs (pre-miRNAs) and transported from nucleus into the cytoplasm. In the cytoplasm, the pre-miRNAs are cleaved by RNase III-type endonuclease Dicer, and then are further processed into single stranded mature miRNAs. However, the miRNAs alone are not functional until they are incorporated into the miRNA-induced silencing complex (miRISC).
With the recruitment of a group of RNA Binding Proteins (RBPs), miRISCs are delivered to their mRNA targets in order to regulate the expression of these genes (Figure 1) [24, 25].
Studies have shown that microRNAs play an important role in the pathogenesis of diabetes, due to the stability of these regulatory molecules in body fluids and their availability and measurement technology, they can be used as genetic biomarkers in early diagnosis of diabetes they used it.
miRNA role in diabetes mellitus: Pancreatic β-cells and other insulin-targeted tissues express a well-defined set of miRNAs that are widely distributed in human tissues. It has been proven that the expression of miRNAs in beta cells and insulin target tissues is changed in patients with T1D and T2D, which is probably due to the defective function of these tissues under the disease conditions. A notable exception is miR 375, a pancreatic islet-enriched miRNA that regulates the expression of genes involved in hormone secretion and β-cell mass expansion in response to insulin resistance [26, 27]. The miRNA expression profile of β-cells and insulin target tissues is altered in patients with T1D and T2D, which likely contributes to the dysfunction of these tissues in the disease state. In a study conducted on healthy and diseased mice, the cells of the islets of Langerhans of diabetic mice contain increased levels of several miRNAs, including miR 21, miR 34a, miR 29, and miR 146a, which have detrimental effects on β-cell function. Changes in the miRNA profile related to diabetes in human tissues have also been reported (Table 1) [28].
| Type of miRNA | Expression rate | Gene targeted | Ref. |
|---|---|---|---|
| miR-15a | increase | VEGFA | [29] |
| miR-200 | Incresase | Fog2 | [30,31] |
| miR-126 | Decrease | ADAM-9 | [32] |
| miR-222 | Increase | CAV-1 | [33,34] |
| miR-375 | Increase | Glucose expression | [35] |
| regulation |
Table 1. The most important miRNAs that can be used as biomarkers of diabetes mellitus type- 2.
Functional role of miRNAs in diabetes disease: In addition to regulating gene expression inside the producing cells, several miRNAs are found in blood and other body fluids in association with proteins, microvesicles or lipoprotein complexes. In vitro studies show that miRNAs delivered by exosomes or high-density lipoprotein (HDL) can be actively transferred to recipient cells [36-40]. These observations increase the possibility of the involvement of miRNAs in connection with cellular processes.
Extracellular miRNAs (circulating in the blood) are highly stable and resistant to treatment with ribonucleases, freeze/ thaw cycles, and other severe experimental conditions [41- 44].
Extracellular miRNAs have several additional advantages as potential biomarkers: they are readily available not only in blood but also in other biological fluids (such as urine, saliva, amniotic fluid, and breast milk). They can be detected by highly sensitive and specific quantitative real-time PCR, and most of them are evolutionarily conserved [45-48].
Application of miRNAs as biomarkers of diabetes
The idea of using miRNAs in blood as biomarkers is relatively new and was first proposed to diagnose different types of cancer and autoimmune disease [49, 50]. Currently, by analyzing the studies conducted on the profile of miRNAs in serum and plasma or blood cells, new approaches have been developed to predict the progression of diabetes mellitus. Zampetaki and colleagues were the first to identify the expression profile of blood miRNAs related to T2D [51].
In their study, they analyzed the blood samples of more than 800 people randomly selected from the population of Bronc (province of Bolzano, Italy) and a subset of five miRNAs including miR-15a, miR-28-3p, miR -29b identified miR-223 and miR-126, which showed dysregulation in 80 participants with pre-T2D or T2D. The serum miRNA content of patients with prediabetes or people newly diagnosed with T2D has also been analyzed by other groups. Kong and colleagues found increased expression of seven diabetes-related miRNAs (miR 9, miR 29a, miR 30d, miR 34a, miR 124a, miR 146a and miR 375) in patients with T2D compared to patients who had prediabetes or were prone to diabetes diagnosed with T2D [52]. However, no differences were observed between subjects with normal glucose tolerance and subjects with prediabetes, indicating that the serum levels of these miRNAs are not suitable for predicting susceptibility to T2D. In a study published in 2012, measured miRNAs in the blood and exosomes of 265 patients with various health conditions associated with metabolic syndrome [53]. They detected the upregulation of miR 27a, miR 150, miR 192, miR 320a and miR 375 in patients with T2D and observed a strong correlation between increased fasting glucose levels and increased miR 27a and miR-320a levels. These pioneering studies demonstrate the potential of miRNAs as biomarkers for T2D. This groundbreaking search shows the future of miRNAs as commercial markers for T2D. After that, various studies have suggested different commercial markers for T2D. After that, various studies have suggested different miRNAs including miR-375, miR-126 and miR-23a as potential commercial markers for the recognition of T2D in the general population [54-56]. In a study conducted in 2014 on miRNA profiles in the blood of diabetics, they found that these miRNAs, mir-222 and mir-126, showed increased and decreased expression, respectively. Treatment of individuals with metformin caused a significant decrease in mir-222 [57]. In 2014, investigated miR-126 expression in 320 diabetic, pre-diabetic and healthy subjects. The expression level of this miRNA in the blood of diabetic people was significantly reduced compared to pre-diabetic people. In these people, after dietary control and exercise, the expression of this miRNA had increased significantly. Also, by examining the biomarker power of this miRNA, they found out that it distinguishes diabetic and healthy people with a power of 82% [58]. In a study conducted [59] on 380 diabetic and healthy subjects, they found that the expression of miR-126 and miR-21 was lower in diabetic subjects than in healthy subjects. Also, by examining the expression of these two genes in people with secondary complications of diabetes such as retinopathy and nephropathy, it was found that the expression of the mentioned genes in these people is reduced compared to diabetic people without complications. Also, in a study conducted [60] in 2019 on blood lymphocyte cells of diabetic people in Iran, they found that mir-15a and mir-126 decreased. On the other hand, the target genes of these miRNAs, namely PRKCB and SP1, were associated with increased expression, which causes an increase in DNA instability and double-strand breaks. These researchers concluded that the reduction of the expression of the mentioned miRNAs plays an important role in the development and progression of diabetes by contributing to the development of double strand breaks. By examining miRNA profiles in prediabetic and diabetic subjects, found that mir-222 and mir-148 were decreased in diabetic subjects and were directly related to HBA1C levels [61]. In 2019, by examining the miR-181b gene expression level in patients with type 2 diabetes in the population of Yazd, showed that there is a significant difference in the expression level of this gene in the healthy and patient groups under study (fold change for healthy people=0.38±0.05 and for prediabetic people fold change=0.12±0.34 [62]. The reduced expression of mir-181-b in people with type 2 diabetes and prediabetes compared to the healthy control group indicates that that mir-181b has the potential to be used as a biomarker in type-2 diabetes. [63] studied the effect of miR-222 and miR-15a expression in pre-diabetic (per- T2D), diabetic and healthy groups. For this purpose, ninety People were selected from Yazd Diabetes Center, which included healthy, pre-T2D and T2D people equally. The level of miRNA expression in plasma samples of the target population was determined by Real-time PCR method. The expression of miR-222 in pre-T2D samples was the significance was increased compared to the control group (P<0.001), while no significant difference was observed in pre-T2D subjects compared to the T2D group (P<0.05). The expression of miR-15a was statistically significant in pre-T2D groups. T2D and T2D had a down-regulation (P>0.05) [63].
Conclusion
Diabetes mellitus is a metabolic disorder, and the number of people suffering from this syndrome is rapidly increasing worldwide, leading to adverse health and socio-economic consequences.
Hence, the discovery of new biomarkers to identify people at risk and thus its proper management is highly needed. The latest trend in biomarker discovery is the search for sensitive biomarkers that can be applied to distinguish diseased individuals from their healthy counterparts and to identify different stages of the disease. Another effective advantage of biomarkers is their availability, so they can be easily obtained from body fluids such as saliva, urine, or blood.
Countless studies have proven that miRNAs are expressed in various tissues and cell types. Furthermore, the role of miRNAs in the regulation of metabolic pathways important for adipose differentiation, energy homeostasis, lipid metabolism, insulin secretion and glucose-induced inflammation has been demonstrated. Therefore, dysregulation of miRNAs expression affects a variety of important cellular functions, including cell cycle regulation, apoptosis, and differentiation, and has a significant effect on health and disease development. The findings described in these papers confirm that miRNAs are novel biomarkers for diabetes. The identification of new biomarkers can help to better understand the pathogenesis events involved in diabetes and also be effective for the diagnosis of T2D in the early stages. Indeed, changes in the level of a subset of these small RNA molecules in body fluids promise new clues for early identification of individuals at risk of developing diabetes and its related complications.
Therefore, their effectiveness in predicting the occurrence of diabetes or its complications should be systematically compared with existing biomarkers. Based on current data, extracellular miRNAs may be able to replace or complement other routine measurements in the future.
References
- Whiting DR, Guariguata L, Weil C, et al. IDF diabetes atlas: Global estimates of the prevalence of diabetes for 2011 and 2030. Diabetes Res Clin Pract. 2011;94(3):311-21.
- Ringborg A, Lindgren P, Martinell M, et al. Prevalence and incidence of Type 2 diabetes and its complications 1996-2003-estimates from a Swedish population‐based study. Diabet Med. 2008;25(10):1178-86.
- Chien HY, Lee TP, Chen CY, et al. Circulating microRNA as a diagnostic marker in populations with type 2 diabetes mellitus and diabetic complications. J Chin Med Assoc. 2015;78(4):204-11.
- Ozcan S. Minireview: Microrna function in pancreatic β cells. J Mol Endocrinol. 2014;28(12):1922-33.
- Hashimoto N, Tanaka T. Role of miRNAs in the pathogenesis and susceptibility of diabetes mellitus. J Hum Genet. 2017;62(2):141-50.
- Natarajan R, Putta S, Kato M. MicroRNAs and diabetic complications. J Cardiovasc Transl Res. 2012;5:413-22.
- Dinesen S, El-Faitarouni A, Dalgaard LT. Circulating microRNAs associated with gestational diabetes mellitus: Useful biomarkers? J Endocrinol. 2023;256(1).
- Saeedi Borujeni MJ, Esfandiary E, Taheripak G, et al. Molecular aspects of diabetes mellitus: Resistin, microRNA, and exosome. J Cell Biochem. 2018;119(2):1257-72.
- Friedman RC, Farh KK-H, Burge CB, et al. Most mammalian mRNAs are conserved targets of microRNAs. Genome Res. 2009;19(1):92-105.
- Kozomara A, Griffiths-Jones S. miRBase: Annotating high confidence microRNAs using deep sequencing data. Nucleic Acids Res. 2014;42(D1):D68-D73.
- Ebert MS, Sharp PA. Roles for microRNAs in conferring robustness to biological processes. Cell. 2012;149(3):515-24.
- Small EM, Olson EN. Pervasive roles of microRNAs in cardiovascular biology. Nat. 2011;469(7330):336-42.
- Park E-Y, Woo Y-M, Park J-H. Polycystic kidney disease and therapeutic approaches. BMB Rep. 2011;44(6):359-68.
- Fernandez-Valverde SL, Taft RJ, Mattick JS. MicroRNAs in β-cell biology, insulin resistance, diabetes and its complications. Diabetes. 2011;60(7):1825.
- Stitt-Cavanagh E, MacLoed L, Kennedy CR. The podocyte in diabetic kidney disease. Sci World J. 2009;9:1127-39.
- Zhang C. MicroRNAs in vascular biology and vascular disease. J Cardiovasc Transl Res. 2010;3:235-40.
- Kim M, Zhang X. The profiling and role of miRNAs in diabetes mellitus. Diabetes Res Clin Pract. 2019;1(1):5.
- Ambros V. The functions of animal microRNAs. Nature. 2004;431(7006):350-5.
- Bartel DP. MicroRNAs: Genomics, biogenesis, mechanism, and function. cell. 2004;116(2):281-97.
- Bartel DP. MicroRNAs: Target recognition and regulatory functions. cell. 2009;136(2):215-33.
- Croce CM. Causes and consequences of microRNA dysregulation in cancer. Nat Rev Genet. 2009;10(10):704-14.
- He L, Hannon GJ. MicroRNAs: Small RNAs with a big role in gene regulation. Nat Rev Genet. 2004;5(7):522-31.
- Zamore PD, Haley B. Ribo-gnome: The big world of small RNAs. Science. 2005;309(5740):1519-24.
- Gregory RI, Yan K-p, Amuthan G, et al. The microprocessor complex mediates the genesis of microRNAs. Nat. 2004;432(7014):235-40.
- Ha M, Kim VN. Regulation of microRNA biogenesis. Nat Rev Mol Cell Biol. 2014;15(8):509-24.
- Poy MN, Hausser J, Trajkovski M, et al. miR-375 maintains normal pancreatic α-and β-cell mass. Proc Natl Acad Sci. 2009;106(14):5813-8.
- Roggli E, Gattesco S, Caille D, et al. Changes in microRNA expression contribute to pancreatic β-cell dysfunction in prediabetic NOD mice. Diabetes. 2012;61(7):1742-51.
- Roggli E, Britan A, Gattesco S, et al. Involvement of microRNAs in the cytotoxic effects exerted by proinflammatory cytokines on pancreatic β-cells. Diabetes. 2010;59(4):978-86.
- Houshmand-Oeregaard A, Schrolkamp M, Kelstrup L, et al. Increased expression of microRNA-15a and microRNA-15b in skeletal muscle from adult offspring of women with diabetes in pregnancy. Hum Mol Genet. 2018;27(10):1763-71.
- Belgardt B-F, Ahmed K, Spranger M, et al. The microRNA- 200 family regulates pancreatic beta cell survival in type 2 diabetes. Nat Med. 2015;21(6):619- 27.
- Yu T, Lu X-J, Li J-Y, et al. Overexpression of miR-429 impairs intestinal barrier function in diabetic mice by down-regulating occludin expression. Cell Tissue Res. 2016;366:341-52.
- Dehghani M, Zarch SMA, Mehrjardi MYV, et al. Evaluation of miR-181b and miR-126-5p expression levels in T2DM patients compared to healthy individuals: Relationship with NF-κB gene expression. Endocrinol Diabetes Nutr. 2020;67(7):454-60.
- Li M, Pan S, Qiu A. Roles of microRNA-221/222 in type 2 diabetic patients with post-menopausal breast cancer. Genet Mol Res. 2016;15(2):10-4238.
- Shi Z, Zhao C, Guo X, et al. Differential expression of microRNAs in omental adipose tissue from gestational diabetes mellitus subjects reveals miR-222 as a regulator of ERα expression in estrogen-induced insulin resistance. Endocrinol. 2014;155(5):1982-90.
- Higuchi C, Nakatsuka A, Eguchi J, et al. Identification of circulating miR-101, miR-375 and miR-802 as biomarkers for type 2 diabetes. Metabol. 2015;64(4):489-97.
- Arroyo JD, Chevillet JR, Kroh EM, et al. Argonaute2 complexes carry a population of circulating microRNAs independent of vesicles in human plasma. Proc Natl Acad Sci. 2011;108(12):5003-8.
- Gibbings DJ, Ciaudo C, Erhardt M, et al. Multivesicular bodies associate with components of miRNA effector complexes and modulate miRNA activity. Nat Cell Biol. 2009;11(9):1143-9.
- Vickers KC, Palmisano BT, Shoucri BM, et al. MicroRNAs are transported in plasma and delivered to recipient cells by high-density lipoproteins. Nat Cell Biol. 2011;13(4):423-33.
- Guay C, Regazzi R. Circulating microRNAs as novel biomarkers for diabetes mellitus. Nat Rev Endocrinol. 2013;9(9):513-21.
- Jo S, Chen J, Xu G, et al. miR-204 controls glucagon-like peptide 1 receptor expression and agonist function. Diabetes. 2018;67(2):256-64.
- Kroh EM, Parkin RK, Mitchell PS, et al. Analysis of circulating microRNA biomarkers in plasma and serum using quantitative reverse transcription-PCR (qRT-PCR). Methods. 2010;50(4):298- 301.
- Mitchell PS, Parkin RK, Kroh EM, et al. Circulating microRNAs as stable blood-based markers for cancer detection. Proc Natl Acad Sci. 2008;105(30):10513-8.
- Finkelstein SD, Sistrunk JW, Malchoff C, et al. A retrospective evaluation of the diagnostic performance of an interdependent pairwise MicroRNA expression analysis with a mutation panel in indeterminate thyroid nodules. Thyroid. 2022;32(11):1362- 71.
- Hocaoglu M, Demirer S, Senturk H, et al. Differential expression of candidate circulating microRNAs in maternal blood leukocytes of the patients with preeclampsia and gestational diabetes mellitus. Pregnancy Hypertens. 2019;17:5-11.
- Gilad S, Meiri E, Yogev Y, et al. Serum microRNAs are promising novel biomarkers. PloS one. 2008;3(9):e3148.
- Keller A, Leidinger P, Bauer A, et al. Toward the blood-borne miRNome of human diseases. Nat methods. 2011;8(10):841-3.
- James‐Allan LB, Rosario FJ, Barner K, et al. Regulation of glucose homeostasis by small extracellular vesicles in normal pregnancy and in gestational diabetes. FASEB J. 2020;34(4):5724-39.
- Griffiths-Jones KABM, Kozomara A, Birgaoanu M. S miRBase: From microRNA sequences to function. Nucleic Acids Res. 2019;47:D155.
- Chen X, Ba Y, Ma L, et al. Characterization of microRNAs in serum: A novel class of biomarkers for diagnosis of cancer and other diseases. Cell Res. 2008;18(10):997-1006.
- Lawrie CH, Gal S, Dunlop HM, et al. Detection of elevated levels of tumour‐associated microRNAs in serum of patients with diffuse large B‐cell lymphoma. Br J Haematol. 2008;141(5):672-5.
- Zampetaki A, Kiechl S, Drozdov I, et al. Plasma microRNA profiling reveals loss of endothelial miR-126 and other microRNAs in type 2 diabetes. Circ Res. 2010;107(6):810-7.
- Dehwah MAS, Xu A, Huang Q. MicroRNAs and type 2 diabetes/obesity. J Genet Genomics. 2012;39(1):11-8.
- Collares CV, Evangelista AF, Xavier DJ, et al. Identifying common and specific microRNAs expressed in peripheral blood mononuclear cell of type 1, type 2, and gestational diabetes mellitus patients. BMC Res Notes. 2013;6:1-15.
- Yang Z, Chen H, Si H, et al. Serum miR-23a, a potential biomarker for diagnosis of pre-diabetes and type 2 diabetes. Acta Diabetol. 2014;51:823-31.
- Zhang T, Li L, Shang Q, et al. Circulating miR-126 is a potential biomarker to predict the onset of type 2 diabetes mellitus in susceptible individuals. Biochem Biophys Res Commun. 2015;463(1-2):60-3.
- Rezk NA, Sabbah NA, Saad MS. Role of MicroRNA 126 in screening, diagnosis, and prognosis of diabetic patients in Egypt. Int Union Biochem Mol Bio. 2016;68(6):452-8.
- Ortega FJ, Mercader JM, Moreno-Navarrete JM, et al. Profiling of circulating microRNAs reveals common microRNAs linked to type 2 diabetes that change with insulin sensitization. Diabetes care. 2014;37(5):1375-83.
- Liu Y, Gao G, Yang C, et al. The role of circulating microRNA-126 (miR- 126): A novel biomarker for screening prediabetes and newly diagnosed type 2 diabetes mellitus. Int J Mol Sci. 2014;15(6):10567-77.
- Olivieri F, Spazzafumo L, Bonafè M, et al. MiR-21-5p and miR-126a-3p levels in plasma and circulating angiogenic cells: Relationship with type 2 diabetes complications. Oncotarget. 2015;6(34):35372.
- Samanian S, Mozdarani H, Behmanesh M, et al. Association of Intrinsic and Induced Genomic Instability in Peripheral Blood Lymphocytes of Type 2 Diabetes Patients with Expression Level of Genes PRKCB and SP1 and microRNAS (miR-126 and miR-15a-3p). Acta Med. 2019;35(2):777-82.
- De Candia P, Spinetti G, Specchia C, et al. A unique plasma microRNA profile defines type 2 diabetes progression. PloS one. 2017;12(12):e0188980.
- Aghaei Zarch SM, Vahidi Mehrjardi MY, Babakhanzadeh E, et al. MiR-181b expression levels as molecular biomarker for type 2 diabetes. J Maz Univ Med. 2019;29(176):195-201.
- Sadeghzadeh S, Dehghani Ashkezari M, Seifati SM, et al. Circulating MiR-15a and MiR-222 as potential biomarkers of type 2 diabetes. Diabetes Metab Syndr Obes. 2020:3461-9.
Indexed at, Google Scholar, Cross Ref
Indexed at, Google Scholar, Cross Ref
Indexed at, Google Scholar, Cross Ref
Indexed at, Google Scholar, Cross Ref
Indexed at, Google Scholar, Cross Ref
Indexed at, Google Scholar, Cross Ref
Indexed at, Google Scholar, Cross Ref
Indexed at, Google Scholar, Cross Ref
Indexed at, Google Scholar, Cross Ref
Indexed at, Google Scholar, Cross Ref
Indexed at, Google Scholar, Cross Ref
Indexed at, Google Scholar, Cross Ref
Indexed at, Google Scholar, Cross Ref
Indexed at, Google Scholar, Cross Ref
Indexed at, Google Scholar, Cross Ref
Indexed at, Google Scholar, Cross Ref
Indexed at, Google Scholar, Cross Ref
Indexed at, Google Scholar, Cross Ref
Indexed at, Google Scholar, Cross Ref
Indexed at, Google Scholar, Cross Ref
Indexed at, Google Scholar, Cross Ref
Indexed at, Google Scholar, Cross Ref
Indexed at, Google Scholar, Cross Ref
Indexed at, Google Scholar, Cross Ref
Indexed at, Google Scholar, Cross Ref
Indexed at, Google Scholar, Cross Ref
Indexed at, Google Scholar, Cross Ref
Indexed at, Google Scholar, Cross Ref
Indexed at, Google Scholar, Cross Ref
Indexed at, Google Scholar, Cross Ref
Indexed at, Google Scholar, Cross Ref
Indexed at, Google Scholar, Cross Ref
Indexed at, Google Scholar, Cross Ref
Indexed at, Google Scholar, Cross Ref
Indexed at, Google Scholar, Cross Ref
Indexed at, Google Scholar, Cross Ref
Indexed at, Google Scholar, Cross Ref
Indexed at, Google Scholar, Cross Ref
Indexed at, Google Scholar, Cross Ref
Indexed at, Google Scholar, Cross Ref
Indexed at, Google Scholar, Cross Ref
Indexed at, Google Scholar, Cross Ref
Indexed at, Google Scholar, Cross Ref
Indexed at, Google Scholar, Cross Ref
Indexed at, Google Scholar, Cross Ref
Indexed at, Google Scholar, Cross Ref
Indexed at, Google Scholar, Cross Ref
Indexed at, Google Scholar, Cross Ref
Indexed at, Google Scholar, Cross Ref
Indexed at, Google Scholar, Cross Ref
Indexed at, Google Scholar, Cross Ref
Indexed at, Google Scholar, Cross Ref
Indexed at, Google Scholar, Cross Ref
Indexed at, Google Scholar, Cross Ref
Indexed at, Google Scholar, Cross Ref
Indexed at, Google Scholar, Cross Ref
Indexed at, Google Scholar, Cross Ref
Indexed at, Google Scholar, Cross Ref
Indexed at, Google Scholar, Cross Ref
Indexed at, Google Scholar, Cross Ref
Indexed at, Google Scholar, Cross Ref