Research Article - Biomedical Research (2018) Volume 29, Issue 16
Breast cancer detection using modified Hough transform
Osama R. Shahin1,2*, Meshrif Alruily3, Mansi Alsmarah1 and Musharraf Alruwaill11Computer Science and Information Department, Faculty of Science & Arts, Jouf University, Saudi Arabia
2Physics and Mathematics Department, Faculty of Engineering, Helwan University, Egypt
3College of Computer and Information Sciences, Jouf University, Saudi Arabia
- *Corresponding Author:
- Osama R. Shahin
Computer Science and Information Department
Faculty of Science & Arts
Jouf University, Saudi Arabia
Accepted on August 06, 2018
DOI: 10.4066/biomedicalresearch.29-18-912
Visit for more related articles at Biomedical ResearchAbstract
Breast cancer is a result of the uncontrolled cells division. These cells may spread from the breast to other parts of the body through blood vessels. Many symptoms were associated to the breast cancer, such as change in color, skin, and shape. In this work, the breast tumor will detect according to its geometric, texture, and gradient features. Firstly, region of interest (ROI) will detected by using the Hough transform. After the location of breast tumor was determined, these locations will cropping from the original mammogram image to detect the actual tumor, then twenty two features will be computed for the ROIs using predefined equations. These features are used to classify the infected tissues from the normal one. A comparison was made between system results of the breast features and reading of an expert’s radiologist to ensure the feasibility and algorithm efficiency.
Keywords
Mammograms, Breast cancer, Hough transform.
Introduction
Breast cancer is where anomalous cells develop in an uncontrolled manner. It is the most widely recognized reason for death among moderately aged ladies. The World Health Organization's International Agency for Research on Cancer in Lyon, France, has evaluated that in excess of million ladies overall kicked the bucket because of breast cancer every year [1].
Mammography helps in early location and it assumes a critical part in cancer treatment and permits a quicker recuperation for a large portion of the patients [2]. There are two sorts of mammography, film and computerized. Computerized mammography is superior to film mammography since radiation beams can be lessened up to half can even now identify breast cancer, while in film mammography the standard radiation beams can't be diminished [3,4]. Keeping in mind the end goal to enhance the biopsy yield proportion, masses must be named amiable or threatening.
Masses with sporadic in shapes are normally threatening, and those joined with radiolucent shapes are favourable [5,6]. In this work the digital mammogram images that were provided from online mammogram database (MIAS database) will be used [7]. Firstly, for each image the location for each suspicious areas will be decided and secondly a number of significant features-geometric, texture, and gradient featureswill be computed [6,8-10].
Tumor Features
There are a lot of features types that describe tumors in image. The function of features is to translate the medical statistics into decisions. Firstly, geometric features describe the geometric properties of the ROI. It is represented as a set of pixels in image [11,12]. Secondly, texture is really very fuzzy concept, these features of attempt to describe grayscale items between adjacent pixels in the image [13]. Finally, gradient is a subsidiary of the image among the neighbourhood respects. The Sobel operator is used to record gradient image as well [14].
Methodology
The suspicious area (ROI) can be easily detected in a picture if the area contains enough background variation. The steps of the proposed can be summarized as follows:
Reading the mammogram image
The images that are used in the algorithm from MIAS database [7], are firstly converted into gray level.
Create gradient image
ROI differs significantly in contrast from the background. Changes in contrast can be detected by different operators such as Sobel, Prewitt, Roberts, Canny, etc. In this work, Sobel detector is used, due to its design simplicity.
Subtracting the original image from the image after filling its gaps
After some of pre-processing process such as image dilates, fill interior gaps; the subtraction of the background from the filling images will be preceded. This process is very essential, to enhance the periphery of the ROI to make it easy to fit it in a circle that made in the following stage using Hough transform.
Detect the circular shape using the Hough transform
Hough transform is used here to surround the tumor area by a circle that closes to the surface dimension of the tumor [15]. The pseudo code of the algorithm can be summarized in Table 1.
| Load image |
| Find the edges in the image |
| For each edge point (xo, yo) |
| For (xo=xo_min; xo ≤ xo_max; xo++) |
| For (yo=yo_min; yo ≤ yo_max; yo++) |
| r=sqrt ((x-xo)^2+(y-yo)^2); |
| Accumulator (r) (xo) (yo)++; //Voting |
| Find the average values in accumulator for (r) (xo) (yo) |
Table 1. Pseudo code for modified Hough transform.
Determine the features for the ROI
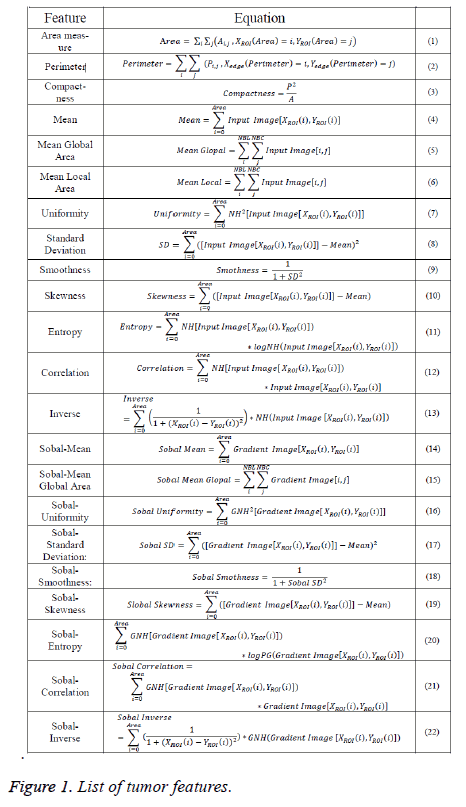
Here, the geometric, texture, and gradient features of ROI will be calculated using equations that predefined in [6,12,13]. Equations that needed to detect tumor features were listed in Figure 1.
Experimental Results
This part introduces the experimental results of the proposed algorithm on the dataset consisting of 322 breast images from MIAS database that covering all cases of masses (malignant and benign) by different shapes, and size. The goal is to test and validate the proposed system presented in this paper in order to select the most suitable system. The total number of malignant samples that used here were 52, while the number for benign was 66 samples; finally normal samples were equal to 204. Table 2 lists the description of all images in the MIAS database and supplies the following significant information [16]. A portion of the tissues are darkened by a splendid area in the focal point of the mammogram. This brilliant locale makes the image be unevenly lit up.
| Fatty tissues | Fatty glandular tissues | Dense tissues | Total | ||
|---|---|---|---|---|---|
| Calcification | B:2 | B:5 | B:5 | B:12 | 25 |
| M:4 | M:4 | M:5 | M:13 | ||
| Circumscribed masses | B:7 | B:6 | B:3 | B:16 | 20 |
| M:2 | M:2 | M:0 | M:4 | ||
| Spiculated masses | B:2 | B:4 | B:6 | B:12 | 21 |
| M:4 | M:3 | M:2 | M:9 | ||
| Other, ill-defined masses | B:4 | B:2 | B:1 | B:8 | 15 |
| M:4 | M:4 | M:1 | M:7 | ||
| Architectural distortion | B:4 | B:3 | B:4 | B:10 | 20 |
| M:2 | M:4 | M:4 | M:10 | ||
| Asymmetry | B:2 | B:3 | B:3 | B:8 | 17 |
| M:3 | M:2 | M:4 | M:9 | ||
| Normal | 66 | 64 | 74 | - | 204 |
| Total | 106 | 104 | 112 | - | 322 |
| B: Benign; M: Malignant. | |||||
Table 2. MIAS database description.
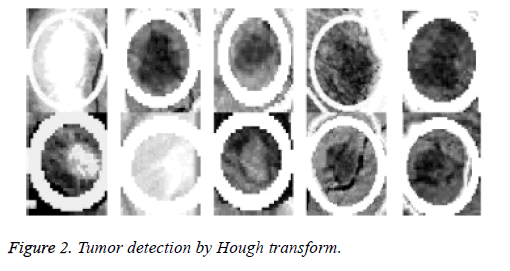
Because the background of the mammogram is dark and the image of tumor masses are lighter than the background of the image, the using of the top-hat transform is required to reduce uneven lighting. This transformation is defined as the difference between the original image and its opening. The opening of the image is the set of front parts of the image that fit a particular structural element. After the location of the tumor area from the entire breast locale the features for these district will be ascertained, as specified before; 22 features will be calculated and will be spared in database which contains 322 row, each row for one image and 22 features extraction that calculated based on equations (1-22). The terminology measurement, i.e. (performance criteria) of the work for system is shown in Table 3. After the location of breast cancer was determined by previous method, these locations will cropping from the original mammogram image to detect the actual tumor as depicted in Figure 2. As a result of breast tumor detection, a 22 features based upon the equations that explained in the methodology section will be calculate that shown in Table 4.
| TN | TP | FP | FN | Sensitivity (%) | Specificity (%) | Accuracy (%) | Precision (%) |
|---|---|---|---|---|---|---|---|
| 164 | 136 | 16 | 6 | 95.77 | 90.9 | 93.17 | 89.47 |
Table 3. Terminology measurement of the algorithm.
| Feature | Maximum | Minimum |
|---|---|---|
| Area | 260 | 75 |
| Perimeter | 3580 | 270 |
| Compactness | 34.554 | 17.456 |
| Mean | 242.802 | 142.133 |
| Mean global area | 9.245 | 1.968 |
| Mean local area | 13.176 | 1.858 |
| Uniformity | 4.993 | 1.177 |
| SD | 32.632 | 12.866 |
| Smoothness | 0.006 | 0 |
| Skewness | 37.466 | -8.203 |
| Entropy | -27.676 | -197.477 |
| Correlation | 569 | 48 |
| Inverse | 0.6446 | 0.001 |
| Sobal mean | 97.459 | 44.597 |
| Sobal mean global | 8.472 | 1.97 |
| Sobal uniformity | 3.583 | 0.005 |
| Sobal SD | 27.639 | 5.948 |
| Sobal smoothness | 0.0538 | 0.001 |
| Sobal skewness | 25.005 | -5.538 |
| Sobal entropy | -20.214 | -250.459 |
| Sobal correlation | 378 | 8 |
| Sobal inverse | 5.846 | 0.001 |
Table 4. Feature extraction range values for cancer tumor.
The results of the computed features by using the equations (2-23) appeared in Table 4. Based upon these features, the classification between the normal tissues and the other infected tissues can be determined by aiding of an expert radiologist.
The performance measures of the proposed algorithm with the other reliable classifiers are depicted in Table 5.
| Author | Methodology | Acc. |
|---|---|---|
| Trbalić et al. [17] | ROIs are obtained using a proficient Otsu's N thresholding and then subjected to pre-processing stage. Then a collection of 32 Zernike moments with dissimilar iterations and orders have been extracted. These moments applied to the neural network classifier. | 90% |
| Liu et al. [18] | The regions of interest (ROIs) are located by using multiple concentric layers (MCL) approach. Then, ROIs are further refined with narrow band based active contour (NBAC), which can enhance the segmentation process. | 92% |
| Xie et al. [19] | A new CAD system depends on the maximum learning machine (ELM). The proposed system contains extraction feature, selection of parameters, and rating. After pre-treatment, areas containing tumor masses are detected. | 95%~96% |
| Proposed | The regions of interest (ROIs) can be detected via modified Hough transform, the find the features of this region by means of predefined equations described in paper, and compare it with normal values with advice of expert radiologist. | 96% |
Table 5. Performance comparison with the other reliable classifiers in terms of accuracy (Acc.).
Conclusion
From the comparison tests that for result shown in Table 5, we see that all features obtained by the proposed algorithm are matching with those features in the previous works. So, the proposed algorithm is able to detect the region of the tumors and determine its locations. In addition to the last benefit it could detect more than one tumor in the same breast region which gives power to the proposed algorithm. As seen from the experimental results and from the performance comparison, the proposed features extraction shows a good result. Thus, it has an ability to implement such breast cancer detection system.
References
- Nixon Mark S, Alberto S Aguado. Feature extraction & image processing for computer vision. Academic Press 2012.
- Sameti M, Rabab Kreidieh W, Jacqueline M-P, Branko P. Image feature extraction in the last screening mammograms prior to detection of breast cancer. IEEE J Selected Topics in Signal Processing 2009; 3: 46-52.
- Madabhushi A, George L. Image analysis and machine learning in digital pathology: Challenges and opportunities. Med Image Analysis 2016; 33: 170-175.
- Mustra M, Mislav G, Rangaraj MR. Re-view of recent advances in segmentation of the breast boundary and the pectoral muscle in mammograms. Med Biol Eng Computing 2016; 54: 1003-1024.
- Qiu Y, Shiju Y, Maxine T, Samuel C, Hong L, Bin Z. Computer-aided classification of mammographic masses using the deep learning technology: a preliminary study. In Med Imaging 2016: Computer-Aided Diagnosis 2016; 9785: 978520.
- Shahin OR. Brain tumor detection using watershed transform. Ann Clin Cytol Pathol 2018; 4: 1096.
- Astley S, Betal D, Cerneaz N, Dance DR, Kok SL, Parker J, Ricketts I, Savage J, Stamatakis E, Taylor P. The mini-MIAS database of mammograms. MAIS database 2018.
- Syed L, Saima J, Manimala S. Telemammography: a novel approach for early detection of breast cancer through wavelets based image processing and machine learning techniques. In Advances in Soft Computing and Machine Learning in Image Processing 2018; 149-183.
- Koundal D, Savita G, Sukhwinder S. Computer aided thyroid nodule detection system using medical ultrasound images. Biomed Signal Processing and Control 2018; 40: 117-130.
- Yeo SY, Romero J, Loper M, Machann J, Black M. Shape estimation of subcutaneous adipose tissue using an articulated statistical shape model. Computer Methods in Biomechanics and Biomedical Engineering: Imaging & Visualization 2018; 6: 51-58.
- d’Anjou M-A. Principles of computed tomography and magnetic resonance imaging. Textbook of Veterinary Diagnostic Radiology-E-Book 2017; 71.
- Dhungel N, Gustavo C, Andrew PB. A deep learning approach for the analysis of masses in mammograms with minimal user intervention. Med Image Analysis 2017; 37: 114-128.
- Mughal B, Nazeer M, Muhammad S, Tanzila S, Amjad R. Extraction of breast border and removal of pectoral muscle in wavelet domain. Biomed Res 2017; 28.
- María Victoria CC, Patricia Rayon V. Circumscribed mass detection in digital mammograms. IEEE 2006.
- Jain L, Devesh M. Application of Hough transform for finding parametric curves. Int J Comput Appl Eng Sci 2011; 1: 100-103.
- Oliveira Júlia EE, Mark OG, Arnaldo de AA, Bastian O, Thomas MD. Toward a standard reference database for computer-aided mammography. In Medical Imaging 2008; 69151Y.
- Šerifović-Trbalić AT, Demirović D, Prljača N, Cattin PC. Classification of benign and malignant masses in breast mammograms MIPRO 2014; 26-30.
- Liu X, Zeng Z. A new automatic mass detection method for breast cancer with false positive reduction. Neurocomputing 2015; 152: 388-402.
- Xie W, Yunsong L, Yide M. Breast mass classification in digital mammography based on extreme learning machine. Neurocomputing 2016; 173: 930-941.