Perspective - Journal of Molecular Oncology Research (2020) Volume 4, Issue 6
Bcl-2 chromosomal translocation t(14;18) in lung cancer tumors and lung biopsy specimens.
Zahra Farsad1, Roya Farzanegan2, Neda Doozandeh2, Mohammad Behgam Shadmehr3, Reza Sheikhnead1*
1Tofigh Daru, Research and Engineering Company, Tehran, Iran
2Tracheal Diseases Research Center, National Research Institute of Tuberculosis and Lung Diseases (NRITLD), Shahid Beheshti University of Medical Sciences, Tehran, Iran
3Lung Transplantation Research Center, National Research Institute of Tuberculosis and Lung Diseases (NRITLD), Shahid Beheshti University of Medical Sciences, Tehran, Iran
- Corresponding Author:
- Dr. Reza Sheikhnejad
Oshida Novel Pharmatech
Tehran
Iran
Tel: +989151496432
E-mail: sheikhnejad@msn.com
Accepted date: October 06, 2020
Citation: Farsad Z, Farzanegan R, Doozandeh N, et al. Bcl-2 chromosomal translocation t(14;18) in lung cancer tumors and lung biopsy specimens. J Mol Oncol Res. 2020;4(6):16-19.
Abstract
BCL-2 gene amplification not only found in non-Hodgkin’s lymphomas but has been also reported in Small Cell Lung Cancers (SCLC). It has been demonstrated that BCL-2, the founding pro-survival BCL-2 family member is highly expressed in 65% of cases. The expression of BCL-2 was generally higher in SCLC cell lines than other solid tumor cell lines including NSCLC. Tumoral tissues were obtained from 45 lung cancer patients, who underwent curative surgery from 2009 to 2011 at Masih Daneshvari Hospital, Tehran/Iran. The tissues were snap-frozen in liquid nitrogen and stored at -80°C until use. Most tumors 60%) were Non-Small Cell Lung Cancer (NSCLC) and 40% diagnosed as Typical Carcinoid (T.C). We also received 25 lung biopsies from patients with non-cancerous lung diseases. All DNAs were extracted using QIAamp DNA mini Kit (QIAGENE cat.51304). The DNA was extracted from protease K digested samples, following the manufacturer’s protocol; and the DNA concentration was measured by Spectrophotometer. To detect the bcl-2 t(14;18) translocation, the PCR-amplifiable mixture contained 5 μl=500 ng dissected DNA; 3 pmol sense primer for the Mbr (5′-GAGAGTTGCTTTACGTGGCC-3′) or for the mcr (5′-CGCTTGACTCCTTTACGTGC-3′); 3 pmol antisense primer JH (5′-ACCTGAGGAGACGGTGACC-3′); 12 μl Taq DNA Polymerase Master Mix RED (Amplicon master mix) up to a final volume of 25 μL. We found no translocation at MBR breakpoint in any tumor specimen. But surprisingly 80% of lung tumors showed distinctive bands (600-700 bp) at MCR breakpoint. The biopsies from non-cancerous patients also showed no MBR breakpoint but interestingly, 77% of smokers exhibited MCR breakpoint while non-smokers had no bcl2 translocation at all. We could suggest that bcl2 translocation may be an early event that might influence cancer cell survival once lung cancer is further advanced to late stages. The result could also suggest that smoking might somehow alter the expression of bcl2 oncogene, most definitely causing bcl2 translocation at MCR breakpoint.
Keywords
Translocation, MBR breakpoint, MCR breakpoint, NSCLC, Carcinoma, BCL-2 oncogene.
Introduction
Apoptosis is a regulated programmed cell death that is triggered in defected cells. This selective cell suicide plays an essential role in numerous physiological and pathological processes [1]. There are two apoptotic pathways-the extrinsic pathway (activated by ligand engagement of cell surface death receptors) and the intrinsic (mitochondrial) pathway. The BCL-2 family of proteins regulate activation of the intrinsic apoptotic pathway in response to cellular stresses such as DNA damage, g-irradiation, oncogene activation etc.
BCL-2 was first identified through chromosomal mapping in follicular lymphoma where constitutive BCL-2 expression is driven from the immunoglobulin locus by the t(14;18) translocation [2-4]. BCL-2 facilitate oncogenesis through cell death resistance [5,6]. Its involvement in t(14;18) in Follicular Lymphoma (FL), has a central role in the inhibition of apoptosis. The t(14;18) translocation causes constitutive overexpression of BCL2 by juxtaposing it to immunoglobulin heavy chain gene enhancer elements. This translocation is found in 20% of DLBCL. In the following years over15 proteins have been added to this family, each containing one or more BCL-2 Homology (BH) domain. Bcl-2-family proteins play central roles in cell death regulation and are capable of regulating diverse cell death mechanisms that encompass apoptosis, necrosis and autophagy [7,8].
In addition to chromosomal translocations as a mechanism for activation of the BCL-2 gene in human malignancies, changes to BCL-2 gene structure or copy number and many additional mechanisms contribute to elevated gene expression, which is estimated to occur in perhaps as many as half of all human cancers. Among the contributing mechanisms are (a) loss of endogenous microRNAs (miRs) that normally repress BCL-2 gene expression which has been documented in chronic lymphocytic leukemia, where the genes encoding miR15 and miR16 become deleted or inactivated by mutations in >70% of these leukemia, and gene hypomethylation, implying altered epigenetic regulation of BCL-2 in some malignancies [9,10].
Altered expression of other antiapoptotic members of the BCL-2 gene family has also been documented in human cancers and leukemias [11-18], although no somatic mutations have been discovered in these genes to date. Loss of miR-29, which represses antiapoptotic family member MCL-1, can occur in chronic lymphocytic leukemia and colon cancers, suggesting at least one responsible mechanism for elevated expression of relatives of BCL-2 [19-21].
BCL-2 gene amplification not only found in non-Hodgkin’s lymphomas but has been also reported in Small Cell Lung Cancers (SCLC) as well [22,23]. It has been demonstrated that BCL-2, the founding pro-survival BCL-2 family member, is highly expressed in 65% of cases [24,25], as is BIM, a proapoptotic BCL-2 family member, which sensitizes SCLC to targeted therapies [26], in particular BH3 mimetics [27]. They evaluated BCL-2 expression in SCLCs compared with other cancers, to confirm prior findings that BCL-2 expression was generally higher in SCLC, and indeed they found SCLC cell lines have higher BCL2 expression on average than other solid tumor cell lines including NSCLC. They also identified two datasets [28,29] with gene expression from SCLC tumors and found significantly (P<0.001) higher BCL-2 expression in SCLC tumors than in NSCLC tumors.
In this study, we attempt to determine the frequency of BCL-2 translocation at MBR and MCR breakpoints in surgically removed lung tumors and biopsy specimens of patients suffering from other types of lung diseases. We will also compare biopsy specimens of smokers versus non-smokers individuals. The experiments are carried out using Rt-PCR technique.
Materials and Methods
Tissue samples
Tumor tissues were obtained from 45 lung cancer patients, who underwent curative surgery from 2009 to 2011 at Masih Daneshvari Hospital, Tehran/Iran. The tissues were snap-frozen in liquid nitrogen and stored at -80°C until use. The histological examination was performed by pathologists at Masih Hospital to determine the tumor histopathologic types. Stained sections under the microscope revealed that most tumors 60% were Non- Small Cell Lung Cancer (NSCLC) and 40% diagnosed as Typical Carcinoid (T.C). The median age of the lung cancer patients was 50.66 (range: 25-75). We also received 25 lung biopsy specimens from patients with non-cancerous lung diseases. The median age of those provided biopsies was 35.7 years (25 to 53).
DNA extraction
All DNAs were extracted using QIAamp DNA mini Kit (QIAGENE cat.51304). The DNA was extracted from protease K digested samples, following the manufacturer’s protocol; and the DNA concentration was measured by Spectrophotometer.
Translocation detection (PCR)
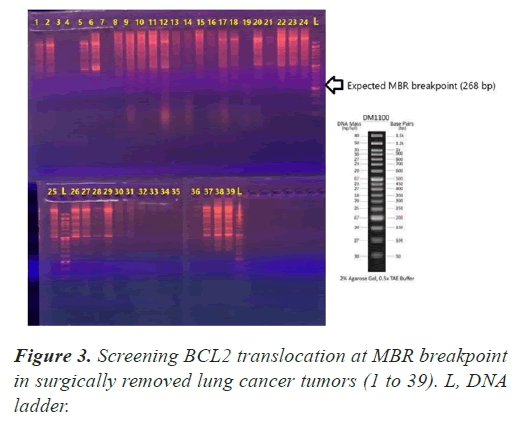
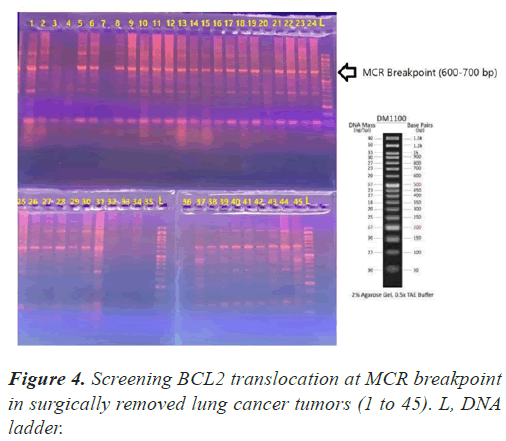
To detect the bcl-2 t(14;18) translocation, the PCR-amplifiable mixture contained 5μl=500ng dissected DNA; 3 pmol sense primer for the Mbr (5′-GAGAGTTGCTTTACGTGGCC-3′) or for the mcr (5′-CGCTTGACTCCTTTACGTGC-3′); 3 pmol antisense primer JH (5′-ACCTGAGGAGACGGTGACC-3′); 12 μl Taq DNA Polymerase Master Mix RED (Amplicon master mix) up to a final volume of 25 μL. The cycling parameter for Mbr and mcr was as follows: initial Denaturation at 95°C for 5 minutes; 35 cycles of 95°C for 1 minute, 58°C for 1 minute, and 72°C for 1 minute; and a final incubation at 72°C for 5 minutes. PCR products for the bcl-2 Mbr and mcr analyses were separated on 1% agarose gels and visualized with autoradiography. Mbr and mcr was analyzed in 39 samples.
Results
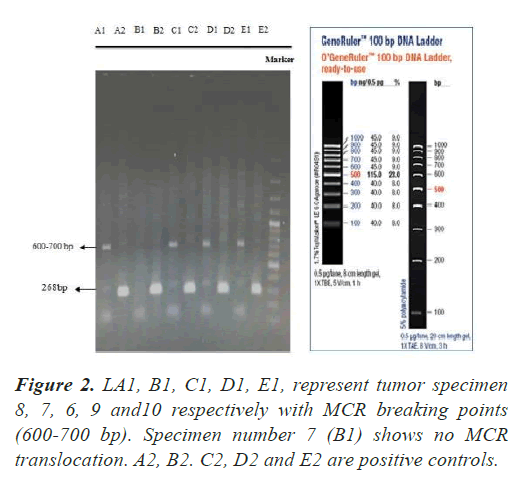
In all our PCR screening tests, we included the extracted DNA from a well-known cancer cell line, DLBCL (Diffuse Large B Cell Lymphoma) for its blc2 translocation at MBR breakpoint. Figure 1 shows a strong band (268 bp) confirming bcl2 translocation at mbr breakpoint next to a tumor specimen with no translocation. The experiment was repeated to show the accuracy of our PCR technique evaluating the extracted DNA from lung tumors and DLBCL cell line using both mbr and mcr primers in determining translocation breakpoints (Figure 2) exhibits bcl2 translocation in our selected tumor specimen and DLBCL at both breakpoints. Screening for bcl2 translocation at MBR breakpoint in lung cancer tumors (1 to 45 as well as some lung biopsies (25 specimens) received from patients with non-cancerous lung diseases. We found no translocation at MBR breakpoint in any tumor specimen (Figure 3). However, when we screened for MCR breakpoints, surprisingly 80% of lung tumor specimens showed distinctive bands (600-700 bp) at MCR breakpoint (Figure 4). Biopsies from non-cancerous patients showed no MBR breakpoint but interestingly, 77% of smokers showed MCR breakpoint while non- smokers had no bcl2 translocation at all (Table 1).
| Biopsy specimens from non-cancerous patients | MCR translocation |
|---|---|
| 10 of 13 smoker biopsy (77%) | Positive |
| 12 of 12 (100%) | Negative |
Table 1: BCL2 translocation (MCR breakpoint) in 25 biopsy specimens of non-cancerous patients.
Discussion and Conclusion
Although we found no report indicating bcl2 translocation in Non-Small Cell Lung Cancer (NSCLC) tumors, we decided to explore if bcl2 plays any role in lung cancers at all. As we collected surgically removed lung tumors, we were hoping to have some SCLC tumors that had been previously reported to have significantly higher BCL-2 expression. However, the histological examination revealed that most tumors 60% were Non- Small Cell Lung Cancer (NSCLC) and 40% diagnosed as Typical Carcinoid (T.C). We found no translocation at MBR breakpoint in any of these tumors. But when we screened for MCR breakpoints, surprisingly 80% of lung tumors showed distinctive bands (600-700 bp) at MCR breakpoint. The biopsy specimens from non-cancerous patients also showed no MBR breakpoint but interestingly, 77% of smokers exhibited MCR breakpoint while non- smokers had no bcl2 translocation at all. The results suggest that bcl2 translocation may be an early event that might influence cancer cell survival once lung cancer is further advanced to late stages. Although we have limited number of biopsy specimens from smoker individual, the result might further suggest that smoking might somehow alter the expression of bcl2 oncogene, most definitely causing bcl2 translocation at MCR breakpoint. Further investigation using large set of specimens is needed to further elaborate on bcl2 role in lung cancer development and prognosis.
References
- Meier P, Finch A, Evan G. Apoptosis in development. Nature. 2000;407:796-01.
- Tsujimoto Y, Finger LR, Yunis J, et al. Cloning of the chromosome breakpoint of neoplastic B cells with the t(14; chromosome translocation. Science. 1984;226:1097-99.
- Bakhshi A, Jensen JP, Goldman P, et al. Cloning the chromosomal breakpoint of t(14; 18) human lymphomas: Clustering around JH on chromosome 14 and near a transcriptional unit on 18. Cell. 1985;41:899-06.
- Cleary ML, Sklar J. Nucleotide sequence of a t (14; chromosomal breakpoint in follicular lymphoma and demonstration of a breakpoint-cluster region near a transcriptionally active locus on chromosome 18. Pro Nat Aca of Sci. 1985;82:7439-43.
- Vaux DL, Cory S, Adams JM. Bcl-2 gene promotes haemopoietic cell survival and cooperates with c-myc to immortalize pre-B cells. Nature. 1988;335:440-42.
- Hockenbery D, Nuñez G, Milliman C, et al. Bcl-2 is an inner mitochondrial membrane protein that blocks programmed cell death. Nature. 1990;348:334-36.
- Cory S, Huang DC, Adams JM. The Bcl-2 family: roles in cell survival and oncogenes is. Oncogene. 2003;22:8590-07.
- Levine B, Kroemer G. Autophagy in the pathogenesis of disease. Cell. 2008;132:27-42.
- Cimmino A, Calin GA, Fabbri M, et al. miR-15 and miR- 16 induce apoptosis by targeting BCL2. Proc Nat Aca Sci. 2005;102:13944-49.
- Hanada M, Delia D, Aiello A, et al. Bcl-2 gene hypomethylation and high-level expression in B-cell chronic lymphocytic leukemia. Blood. 1993;82:1820-28.
- Tron VA, Krajewski S, Klein-Parker H, et al. Immunohistochemical analysis of Bcl-2 protein regulation in cutaneous melanoma. Am J Pathol. 1995;146:643-50.
- Krajewski S, Blomqvist C, Franssila K, et al. Reduced expression of proapoptotic gene BAX is associated with poor response rates to combination chemotherapy and shorter survival in women with metastatic breast adenocarcinoma. Cancer research. 1995;55:4471-78.
- Krajewski S, Chatten J, Hanada M, etal. Immunohistochemical analysis of the Bcl-2 oncoprotein in human neuroblastomas. Comparisons with tumor cell differentiation and N-Myc protein. Laboratory investigation. J Tech Meth Path. 1995;72:42.
- Brousset P, Krajewski S, Meggetto F, et al. Frequent expression of the cell death-inducing gene Bax in Reed-Sternberg cells of Hodgkin’s disease. Blood. 1996;87:2470-75.
- Khanna KK, Wie T, Burrows SR, et al. Expression of p53, Bcl- 2, Bax, Bcl-X2 and c-myc in radiation-induced apoptosis in Burkitt’s lymphoma cells. Cell Death Differ. 1996;3:315-22.
- Krajewska M, Krajewski S, Epstein JI, et al. Immunohistochemical analysis of Bcl-2, Bax, Bcl-X and Mcl-1 expression in prostate cancers. Am J Pathol. 1996;148:1567-76.
- Krajewska M, Kitada S, Winter JN, et al. Bcl-B expression in human epithelial and nonepithelial malignancies. Clin Cancer Res. 2008;14:3011-21.
- Yip KW, Shi W, Pintilie M, et al. Prognostic significance of the Epstein–Barr virus, p53, Bcl-2, and survivin in nasopharyngeal cancer. Clin Cancer Res. 2006;12:5726-32.
- Calin GA, Ferracin M, Cimmino A, et al. A microRNA signature associated with prognosis and progression in chronic lymphocytic leukemia. N Engl J Med. 2005;353:1793-01.
- Cummins JM, He Y, Leary RJ, et al. The colorectal microRNAome. Proc Natl Acad Sci. 2006;103:3687-92.
- Mott JL, Kobayashi S, Bronk SF, et al. Mir-29 regulates Mcl-1 protein expression and apoptosis. Oncogene. 2007;26:6133-40.
- Ikegaki N, Katsumata M, Minna J, et al. Expression of Bcl-2 in small cell lung carcinoma cells. Cancer Res. 1994;54:6-8.
- Monni O, Joensuu H, Franssila K, et al. BCL2 overexpression associated with chromosomalamplification in diffuse large B-cell lymphoma. Blood. 1997;90:1168-74.
- Ben-Ezra JM, Kornstein MJ, Grimes MM, et al. Small cell carcinomas of the lung express the Bcl-2 protein. Am J Pathol. 1994;145:1036-40.
- Jiang SX, Sato Y, Kuwao S, et al. Expression of bcl-2 oncogene protein is prevalent in small cell lung carcinomas. J Pathol. 1995;177:135-38.
- Faber AC, Corcoran RB, Ebi H, et al. BIM expression in treatment-naive cancers predicts responsiveness to kinase inhibitors. Cancer discovery. 2011;1:352-65.
- Faber AC, Farago AF, Costa C, et al. Assessment of ABT- 263 activity across a cancer cell line collection leads to a potent combination therapy for small-cell lung cancer. Proc Natl Acad Sci. 2015;112:1288-96.
- Rohrbeck A, Neukirchen J, Rosskopf M, et al. Gene expression profiling for molecular distinction and characterization of laser captured primary lung cancers. J Transl Med. 2008;6:69.
- BhattacharjeeA, Richards WG, Staunton J, et al. Classification of human lung carcinomas by mRNA expression profiling reveals distinct adenocarcinoma subclasses. Proc Nat Acad Sci. 2001;98:13790-95.