Research Article - Biomedical Research (2017) Volume 28, Issue 8
An evolution based hybrid approach for heart diseases classification and associated risk factors identification
Saman Iftikhar1, Kiran Fatima2, Amjad Rehman1, Abdulaziz S Almazyad4 and Tanzila Saba3*
1College of Computer and Information Systems, Al-Yamamah University, Riyadh, 11512, Saudi Arabia
2Departent of Computer Science, National University of Computer and Emerging Sciences, Islamabad, Pakistan
3College of Computer and Information Sciences, Prince Sultan University Riyadh, 11586 Saudi Arabia
4College of Computer and Information Sciences King Saud University Riyadh Saudi Arabia
- *Corresponding Author:
- Tanzila Saba
College of Computer and Information Sciences
Prince Sultan University Riyadh, 11586, Saudi Arabia
Accepted on December 15, 2016
Abstract
With the advent of voluminous medical database, healthcare analytics in big data have become a major research area. Healthcare analytics are playing an important role in big data analysis issues by predicting valuable information through data mining and machine learning techniques. This prediction helps physicians in making right decisions for successful diagnosis and prognosis of various diseases. In this paper, an evolution based hybrid methodology is used to develop a healthcare analytic model exploiting data mining and machine learning algorithms Support Vector Machine (SVM), Genetic Algorithm (GA) and Particle Swarm Optimization (PSO). The proposed model may assist physicians to diagnose various types of heart diseases and to identify the associated risk factors with high accuracy. The developed model is evaluated with the results reported by the literature algorithms in diagnosing heart diseases by taking the case study of Cleveland heart disease database. A great prospective of conducting this research is to diagnose any disease in less time with less number of factors or symptoms. The proposed healthcare analytic model is capable of reducing the search space significantly while analyzing the big data, therefore less number of computing resources will be consumed.
Keywords
Healthcare analytics, Optimization algorithms, Heart diseases classification.
Introduction
Healthcare information systems are becoming important as they develop relational databases enriched with valuable informative medical data [1-3]. Now-a-days, data mining and machine learning algorithms are being used for analyzing and predicting these huge volume databases. Clinical decision support systems are developed as a result of these algorithms based methodologies. In this way, the risks associated with incorrect diagnostic decisions and the cost associated with wrong clinical medication of patients will be minimized [4,5]. The primary goal of healthcare analytics is to develop different predictive models in various medical domains like heart diseases, cancers, diabetes and other complex diseases [6-8].
Different data mining techniques are being used in healthcare informatics such as: supervised learning technique, unsupervised learning technique and Feature selection techniques [9,10]. These techniques are extensively used in numerous real world applications [11,12]. In supervised learning technique such as classification technique, a model is developed to classify different data classes and then this model is used to predict the class of a feature whose class label is unknown [13]. Training and testing data sets are used to classify the objects in different classes. Another data mining technique known as clustering is an unsupervised learning technique which forms clusters of data objects with unknown class labels on account of some similarity measure [14]. The formed clusters are then used to derive association rules which guide the classification of test instances. Feature selection is a data mining technique used to select a subset of attributes that considered to be most fit for making an optimal diagnosis decision based on some selection criteria.
There are various algorithms in use for supervised learning techniques and unsupervised learning technique. These algorithms are being applied in healthcare analytics [15] with more or less efficacy, such as Support Vector Machines (SVM), Artificial Neural Networks (ANN), decision trees, Bayesian networks, Support Feature Machines (SFM) and regression analysis. This research presents a hybrid approach using a supervised learning model based on a well known classifier SVM and evolutionary optimization techniques (Genetic Algorithm (GA) and Particle Swarm Optimization (PSO) [16,17]. The results are evaluated with algorithms reported in literature and have shown considerably improved accuracy of more than 88%. By using the proposed model for the classification and diagnosis of a disease, big data analysis issues may be resolved to a great extent. This paper is organized in five sections; state of art is presented in section 2, proposed methodology is presented in section 3, experiments and results are discussed in section 4 and conclusion in section 5.
Proposed Methodology
Data collection
The data is collected from heart disease databases available at UCI machine learning data set online repository as the Cleveland heart disease patient's datasets. The data mining problem to be solved is a multiclass problem. The dataset is being segregated into five classes, such as: 0 corresponding to absence of heart disease and 1,2,3,4 corresponding to four different types of heart diseases (Acute Myocardial Infarction (AMI), Percutaneous Coronary Intervention (PCI), Percutaneous Tran luminal Coronary Angioplasty (PTCA) and Coronary Artery Bypass Graft (CABG)). The most imperative 13 attributes and class label are given in Table 1.
| Features | Description | Value |
|---|---|---|
| age | Age | 16-80 |
| sex | Gender | 1: Male 0 : Female |
| cp | Chest Pain Type | 1: typical angina 2: typical type angina 3: non-angina pain 4: asymptomatic |
| trestbps | Trest Blood Pressure | mm Hg on admission to the hospital |
| chol | Serum Cholesterol | (mg/dl) |
| fbs | Fasting Blood Sugar | 0: <120 mg/dl 1: >120 mg/dl |
| restecg | Resting electrographic results | 0: normal 1: having ST-T wave abnormality 2: showing probable or definite left ventricular hypertrophy |
| thalach | Maximum heart rate achieved | |
| exang | Exercise induced angina | 0 = no 1 = yes |
| oldpeak | St depression induced by exercise relative to rest | |
| Slope | Slope of the peak exercise ST segment | 1: unsloping 2: flat 3: downsloping |
| Ca | Number of major vessels colored by floursopy | 0–3 |
| thal | 3: normal 6: fixed defect 7: reversible defect |
|
| Num | Predicted attribute | 0,1,2,3,4 |
Table 1. Data set attributes.
Heart diseases classification through SVM
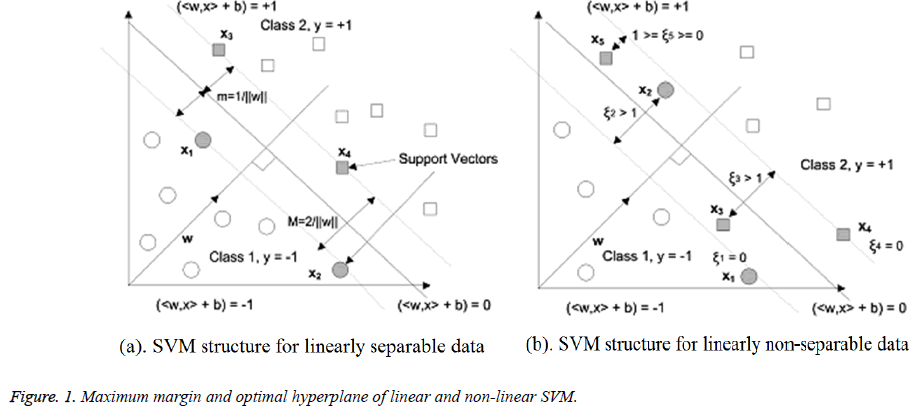
SVMs build a separating hyperplane represented linearly in space of training points. The decision function for classifying linearly separable and non-separable data points with respect to the optimal hyperplane is shown in Figure 1. In this research, experiments are performed to use SVM classifier with three kernel functions and three methods for finding separating hyperplane. Following are the steps of implementation for heart diseases classification through SVM.
1. Load the data set in variable 'data' acquired from UCI repository.
2. Extract the classes in variable ' labels ' mentioned as 0 (no disease) and 1,2,3,4 (other types of heart diseases)
3. Perform training and test sets division through 10-fold cross validation and save the resulting train and test sets in variables 'train' and 'test'
4. Build the SVM structure through Matlab Bioinformatics toolbox builtin function 'svmtrain'
5. SVMStruct = svmtrain(data(train,:),labels(train),'Kernel_Function', kernel_func, 'Method', hyper_method, 'boxconstraint',bx)
6. Classify the test data instances through built in function 'svmclassify' svmclassify(SVMStruct,data(test,:))
7. Evaluate the classifier performance in terms of correct rate (accuracy)
Heart diseases classification through GA-SVM
Genetic Algorithm (GA) is a computational model inspired by evolution and has a basic motivation from the laws of natural selection and genetics. This algorithm preserves significant information through the repetitive application of genetic operations on evolved individuals. In this way, the evolutionary algorithm selects “N best features for classification out of total M features”. Following main steps are performed to implement GA in combination with SVM for heart diseases classification.
1. Generate initial random population of binary strings (chromosomes) to represent presence (1) or absence (0) of features
1. Generate initial random population of binary strings (chromosomes) to represent presence (1) or absence (0) of features
3. Get fitness value (classification accuracy) of random individuals using phenotypes through SVM
4. Perform genetic operation 'tournament selection' to select parent(s) individuals based on fitness value for application of recombination operators crossover and mutation
5. Apply one point crossover and bit-wise uniform mutation on selected parents individuals
6. Remove repetitive individuals from evolved population
.7. Repeat step 2 to step 6 till stagnation (no improvement in fitness value) for a specified number of generations
8. Return optimal feature subset with highest fitness value
Heart diseases classification through PSO-SVM
Particle Swarm Optimization (PSO) algorithm is another population-based optimization algorithm inspired from the social behavior of bird flocking or fish schooling. PSO is widely used in artificial intelligence applications for solving different optimization problems. In PSO, all the potential candidate solutions, called particles, trace the current optimum particles and move through the search space in order to find the global best solution.
From the heart diseases data set, each particle (data point) is taken as a point in N dimensional problem space. Every particle holds a position in terms of its coordinates in search space which is to be updated with a velocity. Each particle's position and velocity is updated in coordination with the best particle based on fitness value (classification accuracy) that has attained through SVM so far by that particle.
Experiments and Results
Parameters setting
In this research, the parameters of SVM kernel functions are tuned in order to get optimal results. Three kernel functions (Linear, Polynomial and Gaussian Radial Basis Function-RBF) of SVM classifier are explored. Three methods for finding the separating hyperplane of SVM are explored including Quadratic Programming (QP) algorithm, Least Square (LS) algorithm and Sequential Minimal Optimization (SMO) algorithm. The values of cost parameter or box constraint, order of polynomial function and RBF scaling parameter sigma are found through grid search algorithm.
The parameters setting used for GA in our experiments is as follows:
No. of Generations = 50, Population Size = 50, Chromosome Length = 13,
Selection mechanism - Tournament (size = 7), Mutation rate = 0.01, Crossover rate = 0.8.
The parameters setting used for PSO algorithm in our experiments is as follows:
No. of generations = 50, Population Size = 50, Particle Length = 13.
Classification results
In this research, a number of experiments are performed in order to explore the optimal combination of SVM parameters in simple mode and in hybrid mode with GA and PSO. The four test scenarios with highest classification results are only given and explained.
In first two scenarios, SVM is explored in simple mode for classification without feature selection.
Scenario 1: SVM with Least Square hyperplane finding method and Linear kernel function (SVM-LS-Linear) using 10- fold cross validation for training and testing divisions achieved mean accuracy of 87.56% over 5 States of heart disease by using 13 attributes (symptoms).
• Mean Accuracy over 5 States: 87.56%
For type 0 - Individual Accuracy: 83.17%
For type 1 - Individual Accuracy: 81.84%
For type 2 - Individual Accuracy: 88.23%
For type 3 - Individual Accuracy: 88.89%
For type 4 - Individual Accuracy: 95.64%
Scenario 2: SVM with Sequential Minimal Optimization hyperplane finding method and RBF kernel function (SVM-SMO-RBF) having = 12 using 10-fold cross validation for training and testing divisions has achieved mean accuracy of 87.95% over 5 States of heart disease by using 13 attributes (symptoms).
• Mean Accuracy over 5 States: 87.95%
For type 0 - Individual Accuracy: 85.83%
For type 1 - Individual Accuracy: 81.84%
For type 2 - Individual Accuracy: 88.24%
For type 3 - Individual Accuracy: 88.23%
For type 4 - Individual Accuracy: 95.63%
In other two scenarios, SVM is explored in hybrid mode for classification with feature selection using GA and PSO algorithm.
Scenario 3: The GA is used for optimal feature selection where the quality (fitness) of a feature set is evaluated through SVM classifier. The highest classification results are obtained when SVM is used with RBF kernel function (=12) and SMO hyperplane calculation method. The mean accuracy over 5 States achieved through GA and SVM hybrid approach is 88.10%.
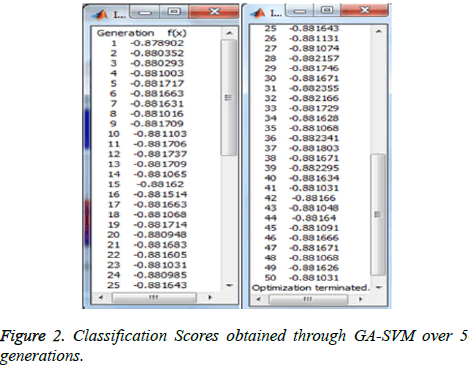
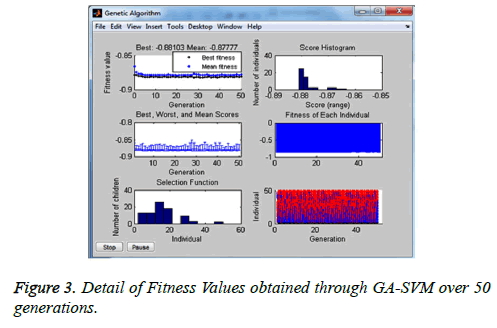
The experiment with GA has shown the following scores (classification accuracy) for 50 generations as in Figure 2 where the 31st generation has given the best fitness value. The detailed graphs of GA individuals and fitness values obtained through GA-SVM over 50 generations as shown in Figure 3. The final chromosome of GA representing the best individual with highest fitness value is shown in Figure 4. This optimal subset of features is showing the set of attributes which are at high risk while diagnosing heart disease.
The reduced feature set obtained through GA includes these attributes: 3 - Chest pain type, 7 - resting electrographic results, 8 - thalach (maximum heart rate achieved) , 9 - exercise induced angina, 11 - slope (the slope of the peak exercise ST segment), 12 - ca (number of major vessels colored by floursopy) and 13 - Thal.
Scenario 4: The PSO algorithm is also employed for the selection of best feature set. The quality (fitness) of a feature set is evaluated through SVM classifier. In this way classification precision is attained by the feature subset (particle) as its final best point. The highest classification accuracy of 88.24% is obtained with RBF kernel function of SVM (Sigma value = 15) and SMO hyperplane calculation method.
The PSO global best particle β (final best point in search space) with high risk features is shown in Figure 5. Digit '1' in the figure represents the high risk features: i.e., 3, 7, 8, 9, 11, 12, 13.
Comparison of heart diseases classification techniques
Overall results of the proposed hybrid approach and other different hybrid and non-hybrid techniques for heart diseases classification and associated risk factors identification are given in Table 2. The classification results achieved by the proposed approach in four scenarios (SVM-LS-Linear, SVMSMO- RBF, GA-SVM-SMO-RBF and PSO-SVM-SMO-RBF) have outperformed the other techniques for heart diseases classification [8-10].
| Classification Algorithms | Accuracy (%) |
| SVM-Linear [18] | 86.62 |
| SVM-Polynomial [19] | 83.9 |
| GA-SVM [19] | 72.55 |
| SFM [20] | 83.31 |
| OCSFM [20] | 86.73 |
| NaïveBayes [18] | 78.93 |
| ANN [18] | 86.04 |
| SVM-LS-Linear (Proposed simple approach) | 87.56 |
| SVM-SMO-RBF (Proposed simple approach) | 87.95 |
| GA-SVM-SMO-RBF (Proposed hybrid approach) | 88.10 |
| PSO-SVM-SMO-RBF (Proposed hybrid approach) |
Table 2. Comparison of classification results of different techniques for multiclass heart disease problem.
Conclusion
In this paper, an evolution based hybrid approach is proposed which exploits SVM classifier, GA and PSO optimization techniques for the classification of multiple states of heart disease. GA and PSO algorithm are used to select less but discriminative features in order to significantly improve SVM classification accuracy. The population-based evolutionary algorithms GA and PSO found the same reduced optimal feature set and best final point respectively. Therefore, the search space for identifying the best solution during heart disease data analysis is reduced that in turn reduced the computing rescores consumption. Moreover, a patient visiting a cardiologist for the examination of a heart disease, he may be examined for the discovered optimal subset of features (symptoms) in less time with less effort. The optimal feature set is obtained with high accuracy and shown high risk associated with the presence of a particular type of heart disease in a patient.
References
- Rad AE, Mohd Rahim MS, Rehman A, Altameem A, Saba T. Evaluation of current dental radiographs segmentation approaches in computer-aided applications. IETE Technical Review 2013; 30: 210-222.
- Norouzi A, Rahim MSM, Saba A, Rada T, Rehman AE, Uddin M. Medical image segmentation methods, algorithms, and applications. IETE Tech Rev 2014; 31.
- Saman I, Sharifullah K, Zahid A, Muhammad K. GenInfoGuard-A Robust and Distortion-Free Watermarking Technique for Genetic Data. PloS one 2015.
- Fatima K, Arooj A, Majeed H. A new texture and shape based technique for improving meningioma classification. Microscopy Res Tech 2014; 77: 862-873.
- Saba T, Rehman A, Sulong G. An intelligent approach to image denoising. J Theor Appl Informa Technol 2010; 17: 32-36.
- Saba T, Almazyad AS, Rehman A. Online versus offline Arabic script classification. Neural Computing Appl 2016; 27: 1797-1804.
- Jadooki S, Mohamad D, Saba T, Almazyad AS, Rehman A. Fused features mining for depth-based hand gesture recognition to classify blind human communication. Neural Comput Appl 2016.
- Saba T, Rehman A, Sulong G. Cursive script segmentation with neural confidence. Int J Innovat Comput Informa Control (IJICIC) 2011; 7: 1-10.
- Saba T, Rehman A. Machine Learning and Script Recognition. Lambert Academic publisher 2012; 37-39.
- Husham A, Alkawaz H, Saba M, Rehman T, Alghamdi AS. Automated nuclei segmentation of malignant using level sets. Microscopy Res Technique 2016.
- Muhsin ZF, Rehman A, Altameem A, Saba A, Uddin M. Improved quadtree image segmentation approach to region information. Imaging Sci J 2014; 62: 56-62.
- Rehman A, Saba T. Neural network for document image preprocessing. Artificial Int Rev 2014; 42: 253-273.
- Rehman A, Saba T. Off-line cursive script recognition: current advances, comparisons and remaining problems. Art Int Rev 2012; 37: 261-268.
- Saba T, Rehman A, Gulong S. Improved statistical features for cursive character recognition. Int J Innovat Comput Informa Control (IJICIC) 2011; 7: 5211-5224.
- Saba T, Al-Zahrani S, Rehman A. Expert system for offline clinical guidelines and treatment. Life Sci J 2012; 9: 2639-2658.
- Rad AE, Rahim MSM, Rehman A, Saba T. Digital dental X-ray database for caries screening. 3D Research 2016; 7: 1-5.
- Younus ZS, Mohamad D, Saba T, Alkawaz MH, Rehman A, Al-Rodhaan M, Al-Dhelaan A. Content-based image retrieval using PSO and k-means clustering algorithm, Arabian J Geosci 2015; 8: 6211-6224.
- Sundar NA, Latha NPP, Chandra R. Performance analysis of classification data mining techniques over heart disease database. Int J Curr Eng Technol 2012; 2: 470-478.
- Bhatia NN, Chow G, Timon SJ, Watts HG. Diagnostic modalities for the evaluation of pediatric back pain: a prospective study. J Pediatr Orthop 2008; 28: 230-233.
- Adeli A, Neshat M. A fuzzy expert system for heart disease diagnosis. Proc Inte Multiconference Eng Comp Sci Hong Kong 2010.