Research Article - Journal of Biochemistry and Biotechnology (2019) Volume 2, Issue 1
Aminoacyl proline synthesis coupled with ATP regeneration using pyruvate phosphate dikinase
Shin Suzuki and Kuniki Kino*
Faculty of Science and Engineering, Department of Applied Chemistry, Waseda University, 3-4-1 Ohkubo, Shinjuku- Ku, Tokyo 169-8555, Japan
- *Corresponding Author:
- Dr Kuniki Kino
Department of Applied Chemistry
Faculty of Science and Engineering
Waseda University
3-4-1 Ohkubo, Shinjuku-ku, Tokyo 169-8555, Japan
Tel: ++81-3-5286-3211
Fax: +81-3-3232-3889
E-mail: kkino@waseda.jp
Accepted date: April 08, 2019
Citation: Shin Suzuki, Kuniki Kino. Aminoacyl proline synthesis coupled with ATP regeneration using pyruvate phosphate dikinase. J Biochem Biotech 2019;2(1):19-23.
Abstract
Aminoacyl prolines (Xaa-Pro) are valuable materials because they exhibit antihypertensive and hypoglycemic activities. We had previously developed a novel method of amide bond formation using the adenylation domain of nonribosomal peptide synthetase that enabled us to synthesize Xaa-Pro enzymatically. However, this method has two significant issues - the supply of ATP for amino acid activation and inhibition of the adenylation reaction caused by pyrophosphate released from the reaction. To address both of these issues, we focused on pyruvate phosphate dikinase (PPDK), which converts AMP, pyrophosphate, and phosphoenolpyruvate to ATP, phosphate, and pyruvate. We constructed an ATP regeneration system using AaPPDK, a PPDK from Acetobacter aceti NBRC 14818T, and applied this system to L-Trp-L-Pro synthesis by the adenylation domain of tyrocidine synthetase A (TycA-A). Using this system, L-Trp-L-Pro production from 1 mM ATP increased to 1.83 mM (2.7-fold), and 0.94 mM L-Trp-L-Pro was synthesized from 1 mM AMP. Moreover, optimization of the enzyme concentrations increased L-Trp-L-Pro production supplemented with 1 mM ATP or AMP to 3.18 mM or 2.19 mM, respectively. AaPPDK successfully reduced ATP addition by converting AMP to ATP and increased the reaction rate by removing PPi. Therefore, we established an efficient production method for Xaa-Pro coupled with the ATP regeneration system from AMP employing AaPPDK.
Keywords
ATP regeneration; Pyruvate phosphate dikinase; Aminoacyl proline; Nonribosomal peptide synthetase; Adenylation domain
Introduction
Aminoacyl prolines (Xaa-Pro) are dipeptides that contain a proline residue at the C-terminus. These dipeptides exhibit various physiological activities, including antihypertensive activity based on inhibition of angiotensin I-converting enzyme [1,2], hypoglycemic activity based on inhibition of dipeptidyl peptidase IV [3,4], and taste [5]. Xaa-Pro possess great potential for use as pharmaceuticals or functional food additives because of their useful bioactivities.
Various methods of peptide bond formation have been developed to date [6]. Solid-phase peptide synthesis (SPPS) [7] is the most typical chemical method and is well established, produces high yields, and can be used to synthesize a wide range of peptides including Xaa-Pro. However, chemical methods have several drawbacks, including a requirement for a condensation reagent [8], protection and deprotection procedure, and equal amount of byproduct per desirable peptide. Several enzymatic or chemoenzymatic methods were also developed, including reverse reaction of peptidases or proteases [9,10], aminolysis reaction by hydrolases [11,12], and ATP-dependent ligases [13-15]. Although various kinds of dipeptides can be catalytically synthesized by diverse enzymes, Xaa-Pro could not be synthesized by any conventional enzymes.
Nonribosomal peptide synthetase (NRPS) is another enzyme that catalyzes peptide bond formation and is involved in the synthesis of secondary peptide metabolites in bacteria or fungi [16]. The adenylation domain (A domain), one of the essential domains of NRPS, plays a critical role in recognition and activation of amino acid substrates incorporated into a peptide chain [16,17]. Peptide bond formation was reported not only with the NRPS system in its entirety, but also with the truncated A domain alone [18]. Recently, we developed a novel method for catalyzing amide bond formation using the A domain of NRPS [19,20]. In this reaction, the A domain initially activates the amino acid substrate using ATP to form an aminoacyl adenylate intermediate and pyrophosphate (PPi). Following a nucleophilic acyl substitution reaction with various nucleophilic amines, aminoacyl-N-alkylamide is formed with releasing AMP thereafter. This method enabled us to synthesize Xaa-Pro using proline as a nucleophile. However, there are two significant issues in this reaction mechanism and they have been mentioned below.
The first issue is a supply of ATP for the activation of amino acids. The industrial utilization of this mechanism is limited due to the high cost of ATP. To address this issue, an ATP regeneration system from AMP is advantageous and several methods employing this technique have been reported. For example, one method involved the conjugation of polyphosphate-AMP phosphotransferase (PAP) and adenylate kinase [21]. Another method employed conjugation of PAP and polyphosphate kinase 1 [22]. In addition, we previously focused on a class III polyphosphate kinase 2, which had been reported to phosphorylate both AMP and ADP using polyphosphate [23] and constructed a simpler system than the one mentioned above [24]. The second issue is that inhibition of adenylation reaction caused by accumulation of PPi released from the reaction. It is well known that adenylation catalyzed by NRPS A domain is reversible, and the reverse reaction proceeds under excess PPi [25]. In previous research, we overcame this issue by supplementation of inorganic pyrophosphatase, which hydrolyzes PPi to phosphate (Pi) [26] for improved reaction rate and productivity [24]. Although each issue could be solved individually, multiple enzymes were required to address both issues collectively.
We focused on pyruvate phosphate dikinase (PPDK) as the essential enzyme for the system, as it had several ideal characteristics. PPDK catalyzes conversion from AMP, PPi, and phosphoenolpyruvate (PEP) to ATP, Pi, and pyruvate (Pyr). This unique reaction would accomplish both ATP regeneration from AMP and removal of PPi by a single enzyme. PPDK could be used in the production process; however, there have only been reports of bioluminescent assays based on luciferase to date [27-29]. In this study, we introduced PPDK to Xaa-Pro synthesis by an A domain of NRPS. Therefore, we established a simpler system for Xaa-Pro production with reduced ATP addition and improved productivity.
Materials and Methods
Chemicals
All chemicals used in this work were reagent-grade and obtained from Fujifilm Wako Pure Chemical (Osaka, Japan), Kanto Chemical (Tokyo, Japan), and Sigma-Aldrich (St. Louis, MO, USA).
Plasmid construction
The plasmid carrying the gene encoding the A domain of tyrocidine synthetase A (pET/TycA-A) was used as previously described [19]. The gene encoding PPDK from Acetobacter aceti NBRC 14818T (AaPPDK) was amplified from genomic DNA using a specific primer pair (5´-ACC CAT ATG ACG AAA TGG GTT TAC AGC TTC G-3´ for forward and 5´- AAC GAA TTC GAT GAG GCT TTT GCC TTT TTA GC-3´ for reverse) and then inserted into the NdeI and EcoRI sites of pET-21a(+). The resulting plasmid was designated as pET/ AaPPDK.
Recombinant protein expression and purification
Escherichia coli BL21(DE3) cells were transformed with each of the expression plasmids. The recombinant E. coli were grown in Luria-Bertani medium (10 g/L Bacto tryptone, 5 g/L Bacto yeast extract and 10 g/L NaCl) containing 50 µg/mL ampicillin and 100 µM isopropyl-Ã-D-thiogalactopyranoside (IPTG) at 25°C for 18 h. The cells were harvested by centrifugation (4°C, 3,000 à g, 10 min) and resuspended in binding buffer for purification. The cells were disrupted by sonication and centrifuged (4°C, 20,000 à g, 30 min). The resulting supernatants were used for enzyme purification with a HisTrap HP 5 mL column (GE Healthcare, Chicago, IL, USA) equipped ÃKTA prime plus (GE Healthcare). The eluates of each enzyme were desalted using a PD-10 column (GE Healthcare) and eluted in 50 mM Tris-HCl buffer (pH 8.0). The expression and purification of the enzymes were confirmed by SDS-PAGE analysis.
Assay of AaPPDK activity
ATP-producing and ATP-consuming reactions by purified AaPPDK were measured. For the ATP-producing reaction, the reaction mixture contained 1 mM AMP, 1 mM PEP, 1 mM PPi, 10 mM MgSO4, and 0.1 mg/mL AaPPDK in 50 mM Tris-HCl buffer (pH 8.0). For the ATP-consuming reaction, the reaction mixture contained 1 mM ATP, 1 mM Pyr, 1 mM Na2HPO4, 10 mM MgSO4, and 0.1 mg/mL AaPPDK in 50 mM Tris-HCl buffer (pH 8.0). Each reaction was initiated by the addition of enzyme and performed at 37°C for 120 min. The reaction was stopped by heating at 80°C for 10 min and then centrifuged (4°C, 20,000 à g, 10 min). The supernatant was subsequently analyzed by a LaChrom L-7000 series HPLC system (Hitachi High-Technologies, Tokyo, Japan). A method to quantify ATP, ADP, and AMP was described previously [24].
L-Trp-L-Pro synthesis by purified recombinant enzymes
The reaction mixture contained 10 mM L-Trp, 100 mM L-Pro, 1 mM ATP or AMP and PPi, 20 mM MgSO4, 0.1 mg/mL TycA-A, and 0.1 mg/mL AaPPDK in 50 mM Tris-HCl buffer (pH 8.0). The reaction was initiated by the addition of enzymes and performed at 37°C for 72 h. The reaction was stopped by heating at 80°C for 10 min and then centrifuged (4°C, 20,000 à g, 10 min). The supernatant was subsequently analyzed using a LaChrom L-7000 series HPLC system. A method to quantify L-Trp-L-Pro and L-Trp was described previously [24].
Results
Selection and overexpression of PPDK
In general, PPDK catalyzes the conversion from AMP, PPi, and PEP to ATP, Pi, and Pyr in a reversible manner [30]. For ATP regeneration from AMP, PPDK should favor the ATPproducing reaction over the ATP-consuming reaction. We therefore searched for PPDK satisfying this requirement and focused on a report characterizing PPDK from Acetobacter aceti NCIB 8554 [31]. According to the report, the Km value for AMP is much lower than for ATP, and the Vmax value for the ATP-producing reaction was higher than the ATPconsuming reaction [31]. Therefore, we selected AaPPDK from A. aceti NBRC 14818 , a species identical to A. aceti NCIB 8554. The gene encoding AaPPDK was cloned and overexpressed in E. coli. Upon SDS-PAGE analysis, we determined that recombinant AaPPDK was expressed as a soluble protein and purified homogenously.
PPDK activity of AaPPDK
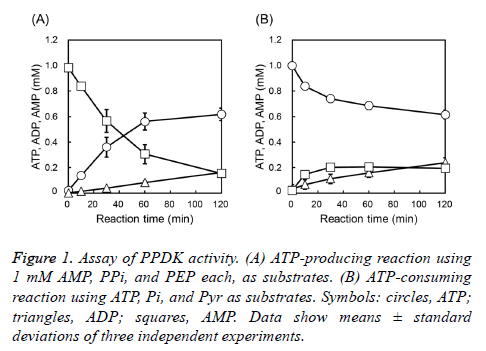
We first investigated the PPDK activity of AaPPDK, as well as the direction of the reaction favored by AaPPDK. The activities towards both the ATP-producing and ATPconsuming reactions were assayed. In the ATP-producing reaction, 0.56 mM of ATP was formed from AMP for 60 min, and moderately increased to 0.62 mM at 120 min (Figure 1A). In contrast, 0.20 mM AMP was formed from ATP for 30 min and AMP was constant after the 30 min-reaction in the ATPconsuming reaction (Figure 1B). These results showed that AaPPDK preferred the ATP-producing reaction and would maintain a high ATP concentration. Therefore, we decided to use AaPPDK for ATP regeneration from AMP.
Figure 1: Assay of PPDK activity. (A) ATP-producing reaction using 1 mM AMP, PPi, and PEP each, as substrates. (B) ATP-consuming reaction using ATP, Pi, and Pyr as substrates. Symbols: circles, ATP; triangles, ADP; squares, AMP. Data show means ± standard deviations of three independent experiments.
L-Trp-L-Pro synthesis coupled with ATP regeneration
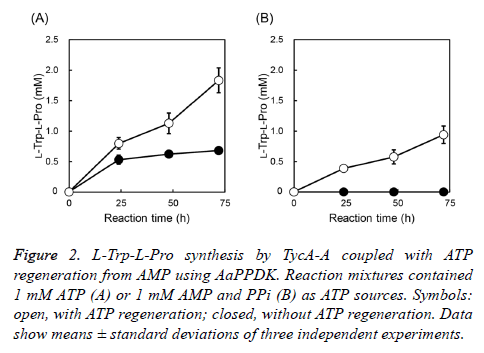
ATP regeneration system using AaPPDK was applied to L-Trp- L-Pro synthesis using TycA-A, which is an A domain of tyrocidine synthetase A from Brevibacillus parabrevis IAM 1031. When 1 mM ATP was supplemented as an ATP source, the reaction rate decreased after 24 h, and 0.68 mM L-Trp-LPro was synthesized after 72 h in the absence of ATP regeneration. By introducing the ATP regeneration, L-Trp-LPro production increased to 1.83 mM (2.7-fold) at 72 h, which exceeded the initial ATP concentration (Figure 2A). Moreover, 0.94 mM L-Trp-L-Pro was synthesized with supplementation of 1 mM AMP in the presence of the ATP regeneration, although no L-Trp-L-Pro was synthesized in the absence of the ATP regeneration (Figure 2B). These results indicated that the ATP regeneration system that employed AaPPDK was functioning effectively.
Figure 2: L-Trp-L-Pro synthesis by TycA-A coupled with ATP regeneration from AMP using AaPPDK. Reaction mixtures contained 1 mM ATP (A) or 1 mM AMP and PPi (B) as ATP sources. Symbols: open, with ATP regeneration; closed, without ATP regeneration. Data show means ± standard deviations of three independent experiments.
Optimization of L-Trp-L-Pro synthesis
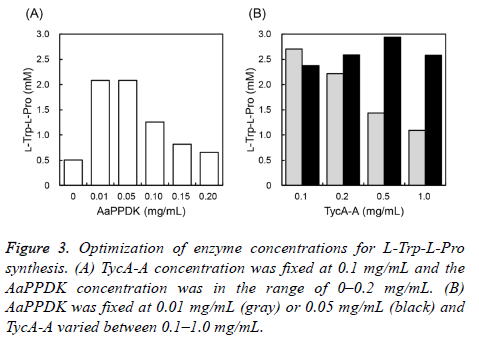
To improve L-Trp-L-Pro productivity, enzyme concentrations were optimized. First, TycA-A was fixed at 0.1 mg/mL and AaPPDK varied between 0?0.2 mg/mL. L-Trp-L-Pro production was the highest when AaPPDK was 0.01 or 0.05 mg/mL and decreased with increasing PPDK concentration (Figure 3A). Next, AaPPDK was fixed at 0.01 or 0.05 mg/mL and TycA-A varied between 0.1?1.0 mg/mL. At 0.01 mg/mL PPDK, L-Trp-L-Pro production decreased with increasing TycA-A concentration. At 0.05 mg/mL PPDK, L-Trp-L-Pro production increased with increasing TycA-A concentration and was the highest at 0.5 mg/mL TycA-A (Figure 3B). From these results, we decided that the optimal conditions were 0.5 mg/mL TycA-A and 0.05 mg/mL AaPPDK.
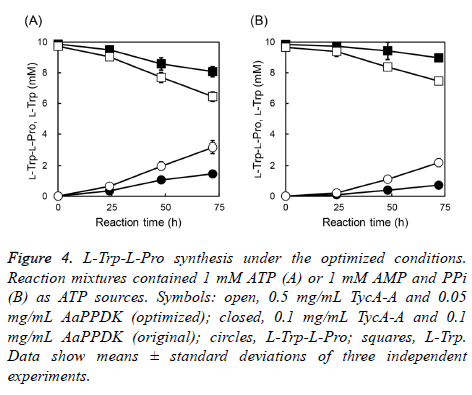
The L-Trp-L-Pro synthesis reaction supplemented with 1 mM ATP or AMP was conducted under the optimized conditions. L-Trp-L-Pro production increased from 1.45 mM to 3.18 mM with ATP and increased from 0.72 mM to 2.19 mM with AMP for 72 h (Figure 4). Optimization delivered better results for LTrp- L-Pro synthesis with respect to the yield and reaction rate.
Figure 4: L-Trp-L-Pro synthesis under the optimized conditions. Reaction mixtures contained 1 mM ATP (A) or 1 mM AMP and PPi (B) as ATP sources. Symbols: open, 0.5 mg/mL TycA-A and 0.05 mg/mL AaPPDK (optimized); closed, 0.1 mg/mL TycA-A and 0.1 mg/mL AaPPDK (original); circles, L-Trp-L-Pro; squares, L-Trp. Data show means ± standard deviations of three independent experiments.
Discussion
In this paper, we constructed an ATP regeneration system from AMP employing AaPPDK in order to address the two issues associated with Xaa-Pro synthesis using A domain of NRPS. We demonstrated the advantages of AaPPDK for L-Trp-L-Pro synthesis as a model reaction.
Although some methods for ATP regeneration from AMP have been reported to date, they required multiple steps and enzymes [21,22]. In contrast, PPDK could convert AMP to ATP in one step, using a single enzyme. This reaction mechanism allowed us to develop a simpler ATP regeneration system than the one mentioned above. AaPPDK from A. aceti NBRC 14818T used in this study favored the ATP-producing reaction over the ATP-consuming reaction (Figure 1). This was an ideal characteristic for the ATP regeneration and successfully functioned in the L-Trp-L-Pro synthesis (Figure 2). Notably, a slight amount of ADP was detected in AaPPDK assay, and this ADP could not be converted to ATP by PPDK; therefore, the ADP formation we observed presents a limitation of the system.
The adenylation reaction in the Xaa-Pro synthesis was inhibited by accumulated PPi [23-25]. AaPPDK uses PPi as a substrate for ATP synthesis, therefore PPi in the coupling reaction might decrease, and the L-Trp-L-Pro production rate improved (Figure 2A). Even though the effect of PPi removal by AaPPDK was weaker than pyrophosphatase, which hydrolyzes PPi to Pi irreversibly, PPi removal was another beneficial effect of AaPPDK on Xaa-Pro synthesis.
Conclusion
In conclusion, we demonstrated two significant effects of AaPPDK on L-Trp-L-Pro synthesis, that is, ATP regeneration from AMP and improving reaction rate by removal of PPi. We previously reported three A domains displaying different substrate specificities and that seven types of Xaa-Pro could be synthesized, including L-Trp-L-Pro. In addition, A domains activating each proteogenic amino acid might exist in nature. Therefore, we speculate that various Xaa-Pro can be synthesized efficiently by introducing the ATP regeneration system from AMP by employing AaPPDK.
Acknowledgements
This work was supported in part by Japan Society for the Promotion of Science (JSPS) KAKENHI [grant number 16K14495]; the Nissin Sugar Scholarship and Research Fund, Tokyo, Japan; and the Waseda University Grant for Special Research Project [grant numbers 2017S-111, 2017B-190, 2018K-251].
References
- Cheung HS, Wang FL, Ondetti MA, et al. Binding of peptide substrates and inhibitors of angiotensin-converting enzyme. Importance of the COOH- terminal dipeptide sequence. J Biol Chem. 1980;255(2):401-407.
- Ichimura T, Hu J, Aita DQ, et al. Angiotensin I-converting enzyme inhibitory activity and insulin secretion stimulative activity of fermented fish sauce. J Biosci Bioeng. 2003;96(5):496-499.
- Hatanaka T, Inoue Y, Arima J, et al. Production of dipeptidyl peptidase IV inhibitory peptides from defatted rice bran. Food Chem. 2012;134(2):797-802.
- Nongonierma AB, Fitzgerald RJ. Inhibition of dipeptidyl peptidase IV (DPP-IV) by tryptophan containing dipeptides. Food Funct. 2013;4(12):1843-9.
- Ishibashi N, Kubo T, Chino M, et al. Taste of Proline-containing Peptides. Agric Biol Chem. 1988;52:95-8.
- Pattabiraman VR, Bode JW. Rethinking amide bond synthesis. Nature. 2011;480(7378):471-479.
- Merrifield RB. Solid phase peptide synthesis. I. The synthesis of a tetrapeptide. J Am Chem Soc. 1963;85(14):2149-2154.
- Valeur E, Bradley M. Amide bond formation: Beyond the myth of coupling reagents. Chem Soc Rev. 2009;38(2):606-631.
- Kumar D, Bhalla TC. Microbial proteases in peptide synthesis: Approaches and applications. Appl Microbiol Biotechnol. 2005;68(6):726-736.
- Yazawa K, Numata K. Recent advances in chemoenzymatic peptide syntheses. Molecules. 2014;19(9):13755-13774.
- Alfonso I, Gotor V. Biocatalytic and biomimetic aminolysis reactions: Useful tools for selective transformations on polyfunctional substrates. Chem Soc Rev. 2004;33:201-9.
- Arima J, Usuki H, Hatanaka T, et al. One-pot synthesis of diverse DL-configuration dipeptides by a Streptomyces D-stereospecific amidohydrolase. Appl Environ Microbiol. 2011;77:8209-18.
- Sato M, Kirimura K, Kino K, et al. D-Amino acid dipeptide production utilizing D-alanine- D- alanine ligases with novel substrate specificity. J Biosci Bioeng. 2005;99:623-628.
- Tabata K, Ikeda H, Hashimoto S. ywfE in Bacillus subtilis codes for a novel enzyme, L-amino acid ligase. J Bacteriol. 2005;187(15):5195-5202.
- Arai T, Arimura Y, Ishikura S, et al. L-amino acid ligase from Pseudomonas syringae producing tabtoxin can be used for enzymatic synthesis of various functional peptides. Appl Environ Microbiol. 2013;79(16):5023-5029.
- Sieber SA, Marahiel MA. Molecular mechanisms underlying nonribosomal peptide synthesis: Approaches to new antibiotics. Chem Rev. 2005;105(2):715-738.
- Schmelz S, Naismith JH. Adenylate-forming enzymes. Curr Opin Struct Biol. 2009;19(6):666-671.
- Dieckmann R, Neuhof T, Pavela-Vrancic M, et al. Dipeptide synthesis by an isolated adenylate-forming domain of non-ribosomal peptide synthetases (NRPS). FEBS Letters. 2001;498(1):42-45.
- Hara R, Suzuki R, Kino K. Hydroxamate-based colorimetric assay to assess amide bond formation by adenylation domain of nonribosomal peptide synthetases. Anal Biochem. 2015;477:89-91.
- Hara R, Hirai K, Suzuki S, et al. A chemoenzymatic process for amide bond formation by an adenylating enzyme-mediated mechanism. Sci Rep. 2018;8(1):2950.
- Resnick SM, Zehnder AJB. In vitro ATP regeneration from polyphosphate and AMP by polyphosphate:AMP phosphotransferase and adenylate kinase from Acinetobacter johnsonii 210A. Appl Environ Microbiol. 2000;66(5):2045-2051.
- Kameda A, Shiba T, Kawazoe Y, et al. A novel ATP regeneration system using polyphosphate-AMP phosphotransferase and polyphosphate kinase. J Biosci Bioeng. 2001;91(6):557-563.
- Motomura K, Hirota R, Okada M, et al. A new subfamily of polyphosphate kinase 2 (class III PPK2) catalyzes both nucleoside monophosphate phosphorylation and nucleoside diphosphate phosphorylation. Appl Environ Microbiol. 2014;80(8):2602-2608.
- Suzuki S, Hara R, Kino K. Production of aminoacyl prolines using the adenylation domain of nonribosomal peptide synthetase with class III polyphosphate kinase 2 mediated ATP regeneration. J Biosci Bioeng. 2018;125(6):644-648.
- Stulberg MP, Novelli GD. Amino acid-activating enzymes: Methods of assay. In Colowick S,Nathan K (Eds). Methods Enzymol. New York. Academic Press. 1962;5(62):703- 707.
- Josse J. Constitutive inorganic pyrophosphatase of Escherichia coli. 1. Purification and catalytic properties. J Biol Chem. 1966;241(9):1938-1947.
- Sakakibara T, Murakami S, Eisaki N, et al. An enzymatic cycling method using pyruvate orthophosphate dikinase and firefly luciferase for the simultaneous determination of ATP and AMP (RNA). Anal Biochem. 1999;268(1):94-101.
- Arakawa H, Karasawa K, Munakata E, et al. Development of bioluminescent pyrophosphate assay using pyruvate phosphate dikinase and its application to single-nucleotide polymorphism analysis. Anal Biochem. 2008;379(1):86-90.
- Karasawa K, Sano Y, Arakawa H. Development of a novel telomerase assay using the PPDK-luciferin-luciferase detection system. Luminescence. 2014;29(1):52-57.
- Chastain CJ, Failing CJ, Manandhar L, et al. Functional evolution of C-4 pyruvate, orthophosphate dikinase. J Exp Bot. 2011;62(9):3083- 3091.
- Schwitzguebel JP, Ettlinger L. Pyruvate, orthophosphate dikinase from Acetobacter aceti. Arch Microbiol. 1979;122:103-8.