Research Article - Biomedical Research (2017) Volume 28, Issue 6
Advanced image segmentation techniques for accurate isolation of abnormality to enhance breast cancer detection in digital mammographs
Vedanarayanan V1* and Nandhitha NM21Research Scholar, Sathyabama University, India
2Department of Electronics and Communication Engineering, Sathyabama University, India
Accepted date: November 12, 2016
Abstract
In recent years, extensive research is carried out in Computer Aided Interpretation of digital mammograms for breast cancer classification. Computer aided Interpretation of digital mammograms involves pre-processing, contrast enhancement, segmentation, appropriate feature extraction and classification. Though considerable research is carried out in developing contrast enhancement and image segmentation techniques, cancer regions could not be isolated and extracted efficiently. Also appropriate features which best describe the cancer characteristics were not found. Hence this work focuses on developing efficient image segmentation techniques for isolating the cancer region and also identifying suitable descriptors for describing the cancer region. Modified Expectation Maximization and modified snake algorithm are developed for isolating the abnormality. Area, Minor Axis Length, Major Axis Length, Perimeter, Orientation, Centroid, Eccentricity, EquivDiameter, Solidity and convex area are the features used for describing abnormality. Back Propagation Network is used for determining the presence and absence of cancer in mammograms. Sensitivity of the proposed techniques is 100%.
Keywords
Digital mammograms, Modified snake algorithm, Modified expectation maximization algorithm, Back propagation network
Introduction
Breast cancer is the most commonly occurring disease in women which proves fatal in most cases. In recent years, various diagnostic techniques are developed to identify breast cancer. These techniques include digital mammograms, Infrared thermographs (IR), Computed Tomography (CT), Positron Emission Tomography (PET) etc. [1]. Of all these techniques, digital mammograms are regarded as golden standard for breast cancer detection [2]. X-ray is passed through the region under observation and is detected on thin films at the other end. When X-Ray passes through cancer cells, it is absorbed and hence it appears as high intensity regions in mammograms. On the other hand, in the absence of cancer X-rays penetrate well and deep, hence that portion of the film appears dark or black. The task of radiologists is to analyse the radiographs and determine the intensity difference to identify cancer regions. Even to an expert radiologist, manual interpretation is difficult for low contrast radiographs. Hence, it is necessary to highlight the region of interest through various contrast enhancement techniques on the mammograms. Ankita et al. have done an extensive survey of different image enhancement methods for digital mammographs [3]. Image segmentation is to be performed to isolate the region of interest. Sukassini et al. had studied different segmentation techniques for digital mammographs in detail [4]. After isolating the region of interest; suitable descriptors are used to describe the cancer region. These features are used to generate exemplars which in turn can be used for training the BPN. Hence the research work aims at developing a suitable image enhancement and segmentation technique for isolating the cancer region completely.
This paper is organised as follows. Section 2 deals with research database and mammograph pre-processing. In section 3, the proposed segmentation techniques are explained. Six layered Back Propagation Network based classifier is discussed in section 4. Conclusion and future work is discussed in Section 5.
Research database and mammograph pre-processing
The steps involved in computer aided analysis of mammograms are mammograph acquisition, mammograph pre-processing, contrast enhancement, segmentation, quantitative characterization and classification [5]. In order to perform the research work a set of 50 mammograms were collected from various scan centres in Chennai, India. Of these mammograms, two are of normal persons and the remaining 48 mammograms depict cancer. Five mammograms which best describe normal and cancer conditions are shown in Figure 1.
From the subjective analysis of these mammograms, it is found that cancer region appears has high intensity pixels in mammograms. Owing to the nature of human physiology few undesirable high intensity regions are also present in mammograms. Hence it is necessary to pre-process the radiographs. In order to reduce the computational complexity, the colour image is converted into grey scale image. After preprocessing the mammographs, contrast is enhanced using Gabor filter.
Advanced image segmentation techniques
On the contrast enhanced images, segmentation techniques are used to isolate the abnormality region [6]. In this work modified expectation Maximization technique and modified snake algorithm are used.
Modified expectation maximization method: Expectation maximization algorithm is widely used when the data set contains missing or hidden values. Most biological datasets have this kind of problem. This is due to the problems in the observations or collection of data’s. All kind of biological analysis is carried on the data’s obtained with limited test only. Expectation maximization algorithm involves in guessing an initial value and increasing the probability of acceptance of that initial parameter [7]. This is done in an iterative way. Maximization step is used to evaluate the parameter and if it is not good then discarded and a new guess is done. In contrast to the conventional EM technique, in improved EM, the mean of the missing value is estimated. This kind of statistical estimation helps in reducing the errors which occur in Markov models. The steps involved in Improved EM are shown below:
• Read the image and convert it to grey scale
• Calculate the upper and lower threshold values of the Enhanced image
• Adjust the mean value based on the threshold values
• Evaluate the no of clusters (E step)
• Update the cluster mean value (M step)
• Based on the clusters, the abnormality is segmented.
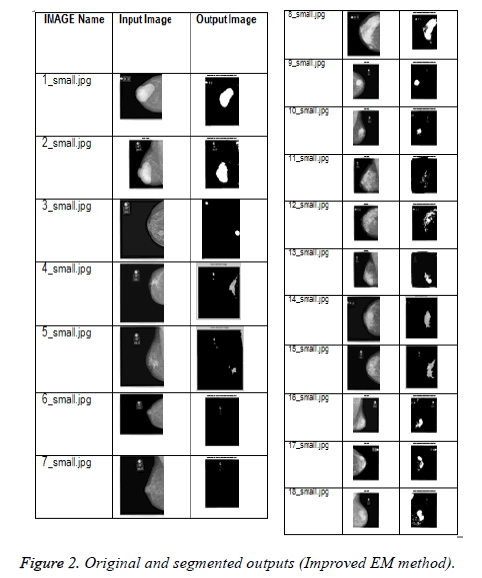
The original and the output images are shown in Figure 2. From the second column of Table 1, it is found that the abnormality regions are isolated accurately.
| Image | Area | Major axis length | Orientation | Perimeter | Centroid | Extrema | Convex area | Solidity | Eccentricity | Equiv diameter | Minor axis length |
|---|---|---|---|---|---|---|---|---|---|---|---|
| 1_small.jpg | 14162 | 183.2055 | 57.6488 | 587 | 209.2732 | 231.5 | 15165 | 0.9339 | 0.8319 | 134.2819 | 101.6591 |
| 2_small.jpg | 13506 | 177.79 | -86.2263 | 665 | 172.1331 | 168.5 | 14880 | 0.9077 | 0.8248 | 131.1349 | 100.5351 |
| 3_small.jpg | 10644 | 756.7651 | -85.3654 | 2230 | 115.5615 | 1.5 | 150076 | 0.0709 | 0.8149 | 116.4146 | 438.6437 |
| 4_small.jpg | 10444 | 760.9611 | -86.0597 | 2230 | 117.6051 | 1.5 | 150076 | 0.0696 | 0.8131 | 115.3157 | 443.0261 |
| 5_small.jpg | 10937 | 765.1262 | -87.6051 | 2220 | 115.4141 | 1.5 | 150076 | 0.0729 | 0.8133 | 118.006 | 445.1632 |
| 6_small.jpg | 10686 | 756.6839 | -84.7703 | 2220 | 116.4376 | 1.5 | 150076 | 0.0712 | 0.8145 | 116.6441 | 439.0256 |
| 7_small.jpg | 10682 | 749.747 | -84.2881 | 2220 | 119.2128 | 1.5 | 150076 | 0.0712 | 0.8075 | 116.6222 | 442.2759 |
| 8_small.jpg | 24447 | 297.6198 | 67.1191 | 2220 | 236.1883 | 1.5 | 150076 | 0.7288 | 0.907 | 176.4281 | 125.3241 |
| 9_small.jpg | 10407 | 769.2389 | -87.0142 | 2220 | 216.8673 | 1.5 | 150076 | 0.0693 | 0.817 | 115.1113 | 443.6133 |
| 10_small.jpg | 10725 | 769.9002 | 88.2337 | 2220 | 217.0283 | 1.5 | 150076 | 0.0715 | 0.8117 | 116.8567 | 449.6466 |
| 11_small.jpg | 1054 | 124.6638 | 8.9493 | 327 | 38.1214 | 1.5 | 150076 | 0.4081 | 0.9746 | 36.6332 | 27.9239 |
| 12_small.jpg | 10872 | 744.0258 | -83.3116 | 2210 | 29.2201 | 1.5 | 150076 | 0.0724 | 0.8063 | 117.6548 | 440.069 |
| 13_small.jpg | 1478 | 51.1846 | 11.1636 | 256 | 230.2463 | 1.5 | 150076 | 0.7549 | 0.4747 | 43.3803 | 45.0494 |
| 14_small.jpg | 745 | 100.215 | -16.5445 | 279 | 25.2148 | 1.5 | 150076 | 0.2884 | 0.9321 | 30.7988 | 36.2867 |
| 15_small.jpg | 10688 | 772.4566 | 89.4908 | 2220 | 116.3498 | 1.5 | 150076 | 0.0712 | 0.8183 | 116.655 | 444.0151 |
| 16_small.jpg | 11196 | 761.9056 | -85.7378 | 2220 | 218.1693 | 1.5 | 150076 | 0.0746 | 0.8121 | 119.3951 | 444.5366 |
| 17_small.jpg | 10991 | 756.1269 | -85.2428 | 2210 | 219.5143 | 1.5 | 150076 | 0.0732 | 0.0732 | 118.297 | 440.3588 |
| 18_small.jpg | 12377 | 772.5146 | -80.797 | 2440 | 189.6908 | 1.5 | 150076 | 0.0825 | 0.7725 | 125.5344 | 490.5579 |
| 19_small.jpg | 10318 | 769.9965 | -85.8297 | 2220 | 212.6296 | 1.5 | 150076 | 0.0688 | 0.8156 | 114.618 | 445.5694 |
| 20_small.jpg | 10461 | 754.2772 | -84.6163 | 2220 | 219.5664 | 1.5 | 150076 | 0.0697 | 0.8135 | 115.4095 | 438.6398 |
| 21_small.jpg | 9784 | 758.9153 | 78.2869 | 2060 | 39.9543 | 1.5 | 150076 | 0.0737 | 0.8505 | 111.6126 | 399.2046 |
| 22_small.jpg | 11170 | 727.2664 | -80.0906 | 2220 | 116.3659 | 1.5 | 150076 | 0.0744 | 0.8008 | 119.2564 | 435.5757 |
| 23_small.jpg | 10715 | 761.108 | -85.4885 | 2220 | 117.6103 | 1.5 | 150076 | 0.0714 | 0.8145 | 116.8022 | 441.5587 |
| 24_small.jpg | 18565 | 691.2519 | 69.0644 | 3220 | 220.7975 | 1.5 | 150076 | 0.1237 | 0.7437 | 153.7455 | 462.1258 |
| 25_small.jpg | 571 | 52.6903 | -7.5053 | 169 | 1079 | 1.5 | 150076 | 0.5292 | 0.8501 | 26.9633 | 27.751 |
Table 1. Quantitative characterization of abnormality isolated by EM method.
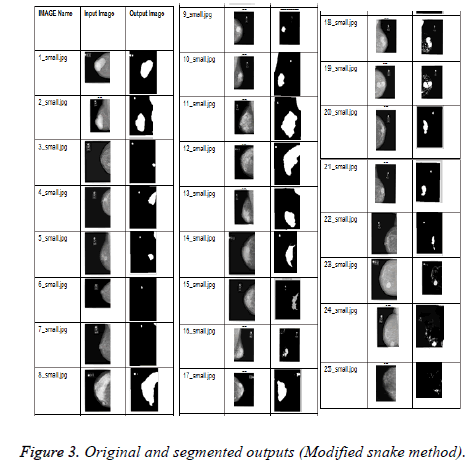
Modified snake algorithm: In order to reduce the computational complexity modified snake algorithm is used for segmenting the abnormality. In general snakes are used for identifying the abnormality of irregular shape. Snake algorithm uses an energy minimising function [8-12]. Snake algorithm is mostly widely used for segmentation of images which contains irregular shape objects. Snake algorithm uses an energy function. This function is a combination of two forces namely internal and external. The internal force is calculated from the shape of the region of interest and the external force is calculated from a higher knowledge about that image. The energy function focuses on three components namely Curvature, Continuity and image gradient. If the edges are smooth then the curvature value should be kept high. In contrast to the conventional snake algorithm, snakes are initially generated by using the histograms. Steps involved in the proposed algorithm are as follows: Initially from the colour image, the mask image is generated. After generating the mask image, it is applied on the original image and the dissimilarity matrix is generated. After a threshold value the snake adjusts itself to reflect the actual abnormality region. The original and the output images are shown in Figure 3. From the figures in the last column of Table 2, it is found that this technique also isolates the abnormality effectively.
| Image | Area | Major Axis length | Orientation | Perimeter | Centroid | Extrema | Convex area | Solidity | Eccentricity | Equiv Diameter | Minor Axis Length |
|---|---|---|---|---|---|---|---|---|---|---|---|
| 1_small.jpg | 2782 | 381.1786 | -83.6799 | 1130 | 57.1736 | 0.5 | 37632 | 0.0739 | 0.8205 | 59.516 | 217.9055 |
| 2_small.jpg | 4390 | 96.0545 | -88.9618 | 269 | 85.8852 | 0.5 | 4564 | 0.9619 | 0.7841 | 74.7631 | 59.6162 |
| 3_small.jpg | 2715 | 379.1589 | -84.98 | 1110 | 58.3153 | 0.5 | 37632 | 0.0721 | 0.815 | 58.7949 | 219.7077 |
| 4_small.jpg | 2744 | 381.3371 | -86.4567 | 1110 | 59.9016 | 0.5 | 37632 | 0.0729 | 0.8108 | 59.1081 | 223.1942 |
| 5_small.jpg | 3343 | 384.9321 | 78.1146 | 1180 | 73.5965 | 0.5 | 37632 | 0.0888 | 0.7686 | 65.2414 | 246.2714 |
| 6_small.jpg | 2702 | 379.6748 | -84.7985 | 1110 | 58.5848 | 0.5 | 37632 | 0.0718 | 0.8158 | 58.654 | 219.5654 |
| 7_small.jpg | 3047 | 385.895 | 87.2854 | 1140 | 68.7939 | 0.5 | 37632 | 0.081 | 0.7837 | 62.2861 | 239.6949 |
| 8_small.jpg | 9478 | 161.303 | 74.8697 | 459 | 119.2717 | 0.5 | 10906 | 0.8691 | 0.8429 | 109.8534 | 86.7846 |
| 9_small.jpg | 4230 | 96.3049 | -70.9107 | 280 | 28.918 | 0.5 | 4501 | 0.9398 | 0.7887 | 73.388 | 59.2063 |
| 10_small.jpg | 5058 | 349.3489 | -64.4684 | 1300 | 71.123 | 0.5 | 37632 | 0.1344 | 0.7307 | 80.2499 | 238.483 |
| 11_small.jpg | 7618 | 124.1984 | 76.8212 | 374 | 96.6533 | 0.5 | 8191 | 0.93 | 0.756 | 98.4862 | 81.2948 |
| 12_small.jpg | 9137 | 160.5529 | 58.741 | 449 | 112.257 | 0.5 | 10191 | 0.8966 | 0.8724 | 107.8591 | 78.4716 |
| 13_small.jpg | 4045 | 91.9671 | -55.8968 | 261 | 106.308 | 0.5 | 4255 | 0.9506 | 0.7849 | 71.7653 | 56.9841 |
| 14_small.jpg | 3723 | 95.6584 | 86.5118 | 254 | 117.6068 | 0.5 | 3860 | 0.9645 | 0.8494 | 68.8496 | 50.4768 |
| 15_small.jpg | 4988 | 105.6116 | 81.4447 | 291 | 136.9493 | 0.5 | 5268 | 0.9468 | 0.8035 | 79.6927 | 62.878 |
| 16_small.jpg | 6377 | 305.4289 | -60.2024 | 1310 | 62.0182 | 0.5 | 37632 | 0.1695 | 0.6957 | 90.108 | 219.4082 |
| 17_small.jpg | 4338 | 96.2214 | 55.7126 | 263 | 103.7937 | 0.5 | 4466 | 0.9713 | 0.7928 | 74.319 | 58.6471 |
| 18_small.jpg | 15128 | 251.6818 | -88.3735 | 780 | 50.2688 | 0.5 | 37632 | 0.402 | 0.7101 | 138.7861 | 177.2085 |
| 19_small.jpg | 11209 | 168.7713 | 89.31 | 448 | 36.3406 | 0.5 | 11309 | 0.9912 | 0.8461 | 119.4644 | 89.9691 |
| 20_small.jpg | 2902 | 372.6751 | -83.9865 | 1110 | 114.1082 | 0.5 | 37632 | 0.0771 | 0.8109 | 60.786 | 218.0975 |
| 21_small.jpg | 2908 | 381.1175 | 86.1361 | 1110 | 110.5519 | 0.5 | 37632 | 0.0773 | 0.8076 | 60.8488 | 224.7676 |
| 22_small.jpg | 2915 | 360.3139 | -80.5463 | 1110 | 58.5674 | 0.5 | 37632 | 0.0775 | 0.7902 | 60.922 | 220.8233 |
| 23_small.jpg | 16137 | 188.9875 | 88.4375 | 521 | 122.0525 | 0.5 | 16260 | 0.9924 | 0.7989 | 143.3397 | 113.6575 |
| 24_small.jpg | 19470 | 240.0864 | 79.177 | 780 | 112.3087 | 0.5 | 37632 | 0.5174 | 0.734 | 157.4483 | 163.0669 |
| 25_small.jpg | 168 | 193.9897 | 0 | 334 | 84.5 | 0.5 | 168 | 1 | 1 | 14.6255 | 1.1547 |
Table 2. Quantitative characterization of abnormality isolated by Modified snake method.
In both cases, abnormality is described by Area, Minor Axis Length, Major Axis Length, Perimeter, Orientation, Centroid, Eccentricity, EquivDiameter, solidity and Convex area. These descriptors are shown in Tables 1 and 2 for improved EM and Modified snake algorithm [13,14]. These parameters are used as input parameters for generating the exemplars to train the neural network.
BPN based classifier: Back propagation network (BPN) is a kind Multi-layer Artificial Neural network which tries to mimic the human decision making process. Back propagation neural network is a supervised training algorithm [15-17]. Back Propagation Network (BPN) uses Gradient descent function which is also known as squashing function to reduce the training error. Back propagation network is much faster than conventional perceptron network. This network uses iterative differentiable activation function so that the error is squashed or reduced to a minimum value. At this stage the desired output matches with the actual output of the network. It has three phases. They are [18] feed forward [19] Error calculation and Error back propagation process [20] weight updating. BPN is used widely for its mathematical simplicity. A six layered Back Propagation Network is used for classification. Number of neurons in the input layer is 11 and one neuron is used at the output layer. Three hidden layers with 22, 11 and 5 neurons are used. “Tansigmoidal” and “purelinear” are the activation functions used at the hidden and the output layers respectively. Learning and momentum parameters are 0.1 and 0.6 respectively. Two different datasets are used for training and testing the neural network. Sensitivity is calculated for exemplars obtained from two segmentation techniques. Relationship between the desired and the actual values are shown in Table 3. From the Table 3, it is found that the sensitivity is 100% for both the segmentation techniques [21].
| S. No | Desired | Actual values for improved EM method | Actual values for improved snake method |
|---|---|---|---|
| 1 | 1 | 1 | 1 |
| 2 | 1 | 1 | 1 |
| 3 | 1 | 1 | 1 |
| 4 | 1 | 1 | 1 |
| 5 | 1 | 1 | 1 |
| 6 | 1 | 1 | 1 |
| 7 | 1 | 1 | 1 |
| 8 | 1 | 1 | 1 |
| 9 | 1 | 1 | 1 |
| 10 | 1 | 1 | 1 |
| 11 | 1 | 1 | 1 |
| 12 | 0 | 0 | 0 |
Table 3. Relationship between the desired and the actual values.
Conclusion and Future work
Two different advanced image segmentation techniques namely improved EM and modified snake algorithm are developed successfully. Region and boundary descriptors are used for quantitatively characterizing the abnormality. These descriptors are used for generating exemplars to train and test the neural network. Based on trained data, the proposed six layered architecture has successfully classified the abnormality from the digital mammographs. Sensitivity of the classifier is 100% for both the two different set of exemplars.
Acknowledgement
The authors acknowledge the various scan centres in Chennai- India which provided the real time images for their research work.
References
- Lee H, Dershaw DD, Kopans D, Evans P, Monsees B, Monticciolo D, Burhenne LW. Breast cancer screening with imaging: recommendations from the society of breast imaging and the ACR on the use of mammography, breast MRI, breast ultrasound, and other technologies for the detection of clinically occult breast cancer. J Am Coll Radiol 2010; 7: 18-27.
- American Cancer Society. Breast cancer facts and figures 2013-2014. Atlanta Ga Am Cancer Soc 2013.
- Ankita P, Singh S, Pandey A. Review: On performance metrics for quantitative evaluation of contrast enhancement in mammograms. Int J Comp Appl 2013; 75: 17.
- Sukassini MP, Velmurugan T. A survey on the analysis of segmentation techniques in mammogram images. Indian J Sci Technol 2015; 8: 22.
- Valarmathi P, Radhakrishna V. Tumor prediction in mammogram using neural network. Glob J Comp sci Technol Neur Artif Intel 2013; 2: 19-24.
- George YM. Breast fine needle tumor classification using neural networks. Int J Comp Sci Iss 2012; 9: 247-256.
- Chang HP, Wei D, Helvie MA, Sahiner B, Adler DD, Goodsitt D, Petrick N. Computer-aided classification of mammographic masses and normal tissue: Linear discriminant analysis in texture feature space. Phys Med Biol 1995; 40: 857-876.
- Jalalian A, Mashohor SB, Mahmud HR, Saripan MI, Ramli AR. Computer-aided detection/diagnosis of breast cancer in mammography and ultrasound: a review. Clin Imaging 2013; 37: 420-426.
- Guo M. A new method for mammographic mass segmentation based on parametric active contour model, Wavelet Analysis and Pattern Recognition (ICWAPR). Int Conf Dat Conf 2015; 27-33.
- Tarver T. Cancer Facts and Figures. American Cancer Society (ACS). J Consu H Intern 2013, 366-367.
- Rao TVN, Govardhan A. Efficient segmentation and classification of mammogram images with fuzzy filtering. Ind J Sci Technol 2015; 8: 15.
- Durgadevi G, Shekhar H. An intelligent classification of breast cancer images. Indian J Sci Technol 2016; 9: 28.
- Sandhya G, Vasumathi D, Raju GT. Mammogram Image segmentation quality enhancement using clustering techniques 2004; 4: 153-158.
- Kamalakannan J, Thirumal T, Vaidhyanathan A, MukeshBhai MD. Study on different classification technique for mammogram image. Circuit, Power and Computing Technologies (ICCPCT). Int Conf Nagercoil 2015; 1-5.
- Priyanka BB, Kulkarni DA. Digital mammography: a review on detection of breast cancer. Int J Adv Res Comp Commun Eng 2016; 5: 386-390.
- Vaidehi K, Subashini TS. A global approach for detecting mass in Digital Mammograms. Int J Adv Res Technol 2012; 1: 6.
- Chaiyakhan K, Kerdprasop N, Kerdprasop K. Feature selection techniques for breast cancer image classification with support vector machine. Proc Int Multi Conf Eng Comp Sci Hong Kong 2016.
- Tang J, Rangayyan RM, Xu J, El Naqa I, Yang Y. Computer-aided detection and diagnosis of breast cancer with mammography: recent advances. IEEE Trans Inf Technol Biomed 2009; 13: 236-251.
- Dougherty G. Medical image processing: techniques and applications. Springe 2011.
- Bovik C. Handbook of image and video processing. Boca Raton CRC Press (2nd Edition.) 2011.
- Jehlol HB. Classification of mammography image using machine learning classifiers and texture features. Int J Innov Res Adv Eng 2015; 9: 56-63.