- Biomedical Research (2011) Volume 22, Issue 3
Active recombinant Reverse Transcriptase Domain of human Hepatitis B Virus Polymerase
Dipendra Raj Pandeya#, Yu Yang# and Seong-Tshool Hong*
Laboratory of Genetics, Department of Microbiology and Immunology, Institute of Medical Science, Chonbuk National University Medical School, Chonju, Chonbuk 561-712, South Korea.
#These authors contributed equally to this work.
- *Corresponding Author:
- Seong-Tshool Hong
Department of Microbiology
Medical school, Chonbuk National University,
Chonju, Chonbuk 561-712, South Korea.
Accepted Date: June 02 2011
Abstract
Hepatitis B virus polymerase plays a critical role during HBV life cycle, and polymerase/reverse transcriptase (RT) activities are critical for HBV-pol during viral replication. To investigate RT do-main of human HBV polymerase, a 5’ end Polyhistidine tagged RT DNA (304-693 amino acids) of HBV-pol was successfully expressed in Escherichia coli. Recombinant RT was purified in native condition employing Ni-NTA affinity column. Purified RT showed a stable reverse transcriptase ac-tivity and a much stronger DNA polymerase activity, compared to RT expressed in rabbit reticulo-cyte lysate coupled transcriptase-translation system. We present a new simplified way of obtaining active RT protein using the Escherichia coli expression and Reticulocyte lysate system. The purified RT was a stable protein and showed a low selective polymerase activity. Computer modeling results also indicated that RT domain banded to nucleotide substrate in a loose mode.
Keywords
Hepatitis B Virus, polymerase, reverse transcriptase, detergent
Introduction
Hepatitis B virus (HBV) infection is a major global public health problem. It is estimated that approximate 1/3 world population has been infected by HBV, and over 350 mil-lion are chronically infected[1,2]. Patients with chronic HBV infection carry a great risk of developing severe liver diseases such as cirrhosis and hepatocyte cellular carcinoma, and these diseases result in a million mortali-ties annually. HBV is a member of the hepadnaviridae family. Despite containing a DNA genome, HBV repli-cates via a reverse transcription process, using the poly-merase encode by its own gene. HBV polymerase (HBV-Pol) consists of three functional domains: (from N-termi-nal to C-terminal) terminal protein (TP) domain, poly-merase/reverse transcriptase domain (RT), and RNase H (RH). TP domain is separated from other two domains by a spacer sequence. During HBV genome replication, HBV progenomic RNA (pgRNA) acts as the template for minus strand of HBV genome DNA replication. The replication is triggered by the protein-priming of TP domain, and then the minus strand DNA synthesis is initiated follow-ing the pgRNA degraded by RH domain of HBV-Pol. The plus strand of HBV genome is synthesized following the nascent minus strand DNA as template.
It is suggested that replication of HBV genome indicated that protein-priming activity, RNase H activity and the two critical activities: RNA-dependent DNA polymerase activity (RDDPa) and DNA-dependent DNA polymerase activity (DDDPa) should be presence in HBV-Pol harmo-niously. Although HBV-Pol which is responsible to RDDPa and DDDPa was documented[3], and then the TP domain and RH domain of human HBV-Pol, which are responsible for protein-priming activity and RNase H ac-tivity respectively, have been proved for decades[4-6]. However, it is still ambiguous whatever, RDDPa and DDDPa are encoded by a specific domain of HBV-Pol or cooperative of the domain of HBV-Pol. Based on the ho-mology of the RT domain of HBV-Pol with known re-verse transcriptase, it is reasonable to believe that the RT domain confer-RDDPa. The difficulties to acquire active recombinant HBV-Pol or RT domain have hampered the characterization of biochemical study of human HBV-Pol. Some groups attempted to express certain domains of hu-man HBV-Pol separately for biochemical study or employ duck HBV polymerase (dHBV-Pol) as research model[7,8]. Expressed TP and RT could form highly sta-ble complex that was active in nucleotide synthesis prim-ing and reverse transcription within insect cells[4]. RDDPa was detected with in insect cells which supplied molecular chaperon such as HSP90 for HBV RT stabiliza-tion, specific binding and possible protein folding assis-tance. However, activities of human HBV RT expressed in rabbit reticulocyte lysate expression system were investigated, and that recombinant HBV-RT exhibited DDDPa lack of RDDPa [9]. Recombinant HBV-RT synthesized in Pichia methanolica also showed DDDPa only [10]. Al-though biochemical research of other domains of human HBV-Pol has been well reported, activities of RT domain of human HBV still need further investigation.
Nowadays, most of approved medications on Chronic Hepatitis B infection (CHB) are nucleotide reverse tran-scriptase inhibitors (NRTIs) such as Lamivudine, Ade-fovir Dipivoxil, Entecavir, Telbivudine and Clevudine, all these NRTIs are targeted on HBV polymerase (HBV-Pol)[1,11]. The spatial relationships between primer-template/reverse transcriptase complex and NRTIs has been well developed [12,13]. To further understand the spatial interaction between RT and template-primer sub-strate complex, a molecular modeling study was also con-ducted employing an existed RT domain modeling tem-plate which conserved motifs and key amino acid residues are coincident with HBV-RT [12]. We have previously described that functional intact human HBV-Pol has been expressed in E. coli without co-expression molecules or in the presence of certain helper chaperons. In this work, stable RDDPa and DDDPa of recombinant HBV-RT ex-pressed in both prokaryotic and eukaryotic expression systems were observed. In terms of modeling result, DDDPa and RDDPa should be presence in human HBV-RT harmoniously, and this result also was coincident with the activity characteristics of recombinant HBV-RT.
Materials and Methods
Computer modeling
A homologous model of reverse transcriptase (protein Data Bank accession no. 1RTD.pdb) as modeling tem-plates was constructed, which covered amino acid resi-dues 319 to 700, comprising the entire RT domain of HBV polymerase. The HBV RT was modeled in a con-formation similar to that of RT domain of HIV poly-merases, with the nucleic acid substrate-binding cleft de-fined by structural elements from the fingers, palm, and thumb sub-domains. A spatial relationship between RT domain and substrate was investigated briefly.
Plasmid construction
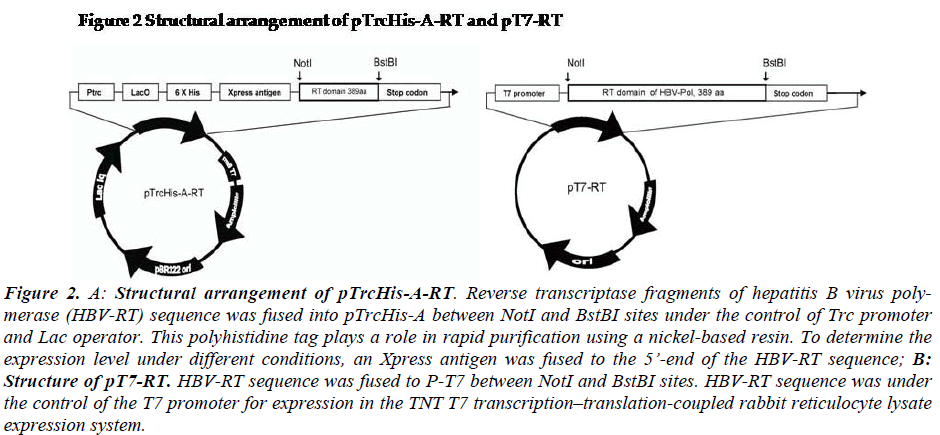
Liver tissue was obtained from a chronic HBsAg carrier who developed hepatocellular carcinoma (HCC) and un-derwent surgical resection (Hospital of Chonbuk National University, Jeonju, South Korea). Briefly, the tumor tis-sues were dissected and immediately cut into small pieces and stored in liquid nitrogen until use. Cellular DNA was isolated from the tissue by SDS-protease K digestion and phenol-chloroform extraction, as described previously (Sambrook). The HBV-RT sequence [spanning 1005-2079 bps,304-693 amino acids] (GenBank accession number AF286594) was amplified with PrimeSTARTM high fidelity polymerase using RTNotIF01: GTT GCG GCC GCT TAA TGG ACT ACT GCC TCA CC and RTBstBIR01: GAA TTC GAA AAT TCC TGA CCG TTG CCG GGC for RT fragments. The vectors were designed as shown in Figure 2 (Fig-2).
For expression of HBV-Pol in E.coli system, the plasmid pTrcHis-A-RT was constructed as showed in Fig-2a. The RT frame was under the control of Trc promoter and Lac operator. His-tag and Xpress antigen were fused at 5’end of RT domain sequence. For expression of HBV-Pol in rabbit reticulocyte lysate system, the plasmid pT7-Pol was constructed as showed in Fig-2b. The HBV-Pol frame was driven by T7-promoter.
The pTrcHis-A-RT and pT7-RT vectors were constructed by the following procedure. RT domain of HBV-Pol full-length sequence was cloned into NotI and BstBI sites of the two expression vector pTrcHis-A (Invitrogen, US). RT domain of HBV-Pol full-length sequence was cloned into the NotI and BstBI sites of the two expression vectors, pTrcHis-A (Invitrogen, US) and pT7 (Promega, US). For expression of HBV-RT in the E.coli system, the plasmid pTrcHis-A-RT was constructed as shown in Fig-2-1. The rhHBV-RT expression was under the control of a Trc pro-moter and a Lac operator. His-tag and Xpress antigens were fused at the 5’ end of the HBV-RT sequence. For expression of rhHBV-RT in a rabbit reticulocyte lysate system, the plasmid pT7-RT was constructed as shown in Fig-1-2. The rhHBV-R expression was driven by the T7-promoter
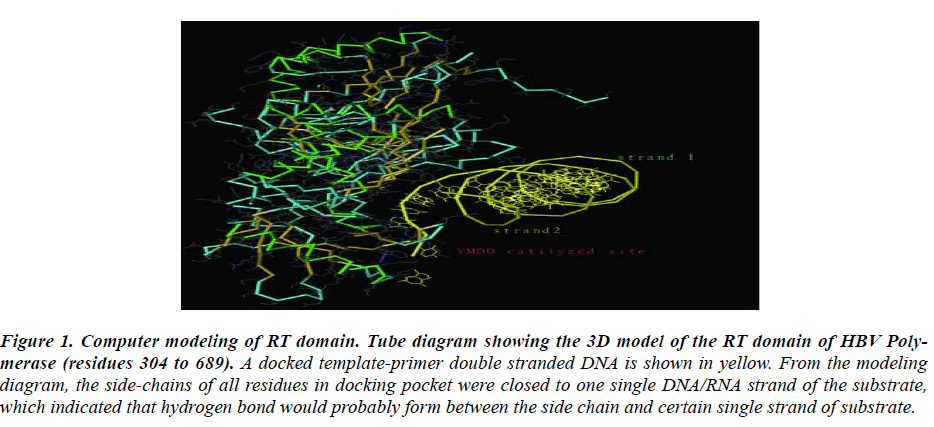
Figure 1: Computer modeling of RT domain. Tube diagram showing the 3D model of the RT domain of HBV Poly-merase (residues 304 to 689). A docked template-primer double stranded DNA is shown in yellow. From the modeling diagram, the side-chains of all residues in docking pocket were closed to one single DNA/RNA strand of the substrate, which indicated that hydrogen bond would probably form between the side chain and certain single strand of substrate.
Figure 2: A: Structural arrangement of pTrcHis-A-RT. Reverse transcriptase fragments of hepatitis B virus poly-merase (HBV-RT) sequence was fused into pTrcHis-A between NotI and BstBI sites under the control of Trc promoter and Lac operator. This polyhistidine tag plays a role in rapid purification using a nickel-based resin. To determine the expression level under different conditions, an Xpress antigen was fused to the 5’-end of the HBV-RT sequence; B: Structure of pT7-RT. HBV-RT sequence was fused to P-T7 between NotI and BstBI sites. HBV-RT sequence was under the control of the T7 promoter for expression in the TNT T7 transcription–translation-coupled rabbit reticulocyte lysate expression system.
E.coli transformation
Plasmid pTrcHis-A-R was chemically transformed in competent DH5α (F-,φ80dlacZΔM15,Δ(lacZYA-argF) U169,deoR,recA1,endA1,hsdR17(rk-,mk+),phoA,supE44,λ-,thi-1,gyrA96,relA1) cells following the manual supplied by Real Biotech Corporation (South Korea).
Expression and purification of histidine-tagged RT do-main
The supernatant was transferred to Ni-NTA resin column (bed volume: 2ml) which had been equilibrated with 16ml equilibration buffer (same components of previous lysis buffer without lysozyme). Resin and lysate supernatant was mixed thoroughly but gently for 60min at 4℃. The resin was washed with 8ml washing buffer (50mM Phos-phate buffer, pH8.0; 0.5mM NaCl; 1%NP-40; 10mM Imi-dazole; 20mM 2-mercaptoethanol; 1X Roche protein in-hibitor cocktail) for 6-8 times. The RT fractions were eluted with 6ml Elution buffer (50mM Phosphate buffer, pH8.0; 0.5mM NaCl; 1%NP-40; 50,100 250mM Imida-zole respectively; 1X Roche protein inhibitor cocktail, dithiothreitol stock solution added in harvest tube first to achieve final concentration to 5mM). RT Elution fractions were harvested and stored at -70℃.
Transformed cells were grown in 100 ml of LB broth until the culture reached an OD600 of 0.6-0.8. Recombinant protein expression was then induced by treatment of the culture with 1 mM Isopropylthiogalactopyranoside (IPTG, Sigma, USA) for 4 h at 37 oC with shaking. Induced cells were harvested by centrifugation at 2,000×g for 20 min at 4 oC. Four volumes of lysis buffer were added to the ob-tained cell pellet. The lysate was incubated at room tem-perature for 30 min and then sonicated for 10×10 sec. The samples were cooled down on ice for 5-10 sec between each sonication. The suspension was centrifuged at 20,000×g for 30 min at 4 oC. The supernatant was trans-ferred to a nickel-based resin (Invitrogen, US) column (bed volume: 2 ml) that had been equilibrated with 16 ml equilibration buffer (same components as the lysis buffer, without lysozyme). The resin and lysate supernatant were mixed thoroughly but gently for 45-60 min at room tem-perature. The resin was washed with 8 ml washing buffer (50 mM phosphate buffer, pH 8.0; 0.5 mM NaCl; 1% NP-40; 10 mM imidazole; 20 mM 2-mercaptoethanol; 1X Roche protein inhibitor cocktail) 6-8 times. The rhHBV-RT fractions were eluted with 6 ml each of E-50, E-75, and E-100 Elution buffer (50 mM phosphate buffer, pH 8.0; 0.5 mM NaCl; 1% NP-40). Roche protein inhibitor cocktail (1X) and dithiothreitol (5 mM) were added to the harvest tubes previously. The concentration of imidazole in E-50, E-75, and E-100 buffers was 50 mM, 75 mM and100 mM, respectively. RhHBV-RT elution fractions were harvested and stored at -70 oC. The protein concen-trations of the purified protein samples were determined by use of a bicinchoninic acid assay using bovine serum albumin (BSA) as a standard.
SDS-PAGE and Western Blot analysis
Total lysate and other purified protein samples were mixed with electrophoresis sample buffer and were dena-tured at 95℃ for 5min. Proteins were separated by SDS-PAGE. Gels were stained with Coomassie Blue. For west-ern blot analysis, protein was electrophoretically trans-ferred to a polyvinylidene difluoride (PVDF) blotting membrane, and membrane was treated with first a 1/4,000 dilution of anti-Xpress antibody followed Goat-anti-mouse conjugated alkaline phosphatase (Invitrogen, US).
Expression of RT domain of HBV polymerase in in vitro expression system
The recombinant plasmid pT7-RT was purified with Qia-gen Midiprep DNA purification kit(Qiagen, Ger). In vitro transcription and translation reactions were per-formed using TNT T7 coupled reticulocyte lysate system (Promega, US). 2 μg of the plasmid DNA template was transcribed and the protein was translated in each 50 ml reaction in the presence or absence of 40 mCi of [35-S]-methionine (1000 Ci/mmol) (Amersham, UK) at 30℃ for 75 minutes[9]. The in vitro translation reaction was stopped by the addition of 0.1 mg/ml cycloheximide for the polymerase activity assay or SDS sample buffer for checking the efficiency of translation. The in vitro trans-lated proteins were separated by 4-12% SDS-PAGE and dried prior to autoradiography.
DNA polymerase activity and reverse transcriptase activ-ity assays
DNA-dependent DNA polymerase activity (DDDP) and reverse transcriptase/RNA dependent DNA polymerase activity (RDDP) were monitored by the synthesis of DNA using poly (dA) · oligo (dT) 12–18 and poly (rA) · oligo (dT) 12–18 as template-primer (Amersham Biosciences Corp.), respectively. The standard enzyme reaction (50 ml) contained 50 mM Tris-Cl pH 7.4, 50 mM KCl, 10mM MgCl2, 1 mM DTT, 0.01% Nonidet P-40, 50 ng of ho-mopolymer template (poly (dA) · oligo (dT) 12–18 for DDDP activity assay and poly (rA) · oligo (dT) 12–18 for RDDP activity assay), and 2 mCi of [α−32P]dTTP (3000 Ci/mmol), ( PerkinElmer, USA). For RDDP activity as-say, RNase inhibitor and RNase free water were em-ployed in the reaction [9,14]. Reactions were started by the addition of 0.5 mg of the purified RT or 5μl products from TNT-T7 coupled reticulocyte lysate system into re-action buffer. The endogenous DNA polymerase activity from the reticulocyte lysate was suppressed by the addi-tion of 60 mM aphidicolin and 1mM NEM. After incuba-tion at 37℃ for 75min, reactions were stopped by the ad-dition of 0.2 mg/ml of protease K in the presence of 0.5% SDS. Incubation was then continued for another 20 min, followed by spotting on Whatman DE81 filter paper. Fil-ters were washed 3 times with 0.5 M Na2HPO4, once with distilled H2O9. Incorporation of radioactivity was deter-mined by liquid scintillation counting in a Packard Tri-Carb Series 2300 liquid scintillation counter.
Results
Molecular Modeling
To understand the spatial interaction relation of RT do-main with nucleotide substrate, a three-dimensional ho-mology model of HBV RT domain was developed based on the HIV RT-DNA crystal structure. The most con-served domains based on the sequence alignment between HIV and HBV-RT 12 and the HIV RT-DNA X-ray struc-ture13 were used to build the HBV RT model. In this study, RT conserved regions surrounding the substrate site was focused. A docked template-primer double stranded DNA was shown in yellow. From the modeling diagram, the side-chains of all residues in docking pocket were closed to a single strand of the substrate, which indicated that hydrogen band would probably form between the side chain and certain single strand of nucleotide sub-strate.
In this study, the HBV-RT domain was encoded by bases 1005 to 2079 of HBV genome and had 389 amino acids.
The RT domain was expressed in two systems: E.coli and reticulocyte lysate system. Then DNA polymerase activity and reverse transcriptase activity from these two protein products were monitored.
Expression, Induction and Purification of HBV-RT in Escherichia coli
To achieve the high expression level, expression level was examined in various conditions. Appropriate induction time is an important factor to obtain enough recombinant RT domain products. Quantity of expressed protein will be varied in different induction time. After induction opti-mization, all the induction culture was carried out at 37℃ for 4h. For HBV-RT purification, HBV-RT fractions were eluted with elution buffers (50, 75, 100mM imidazole). In SDS-PAGE; analysis of western blot also confirmed the purification results (Fig-3b).In this study, a purification
protocol in native condition was applied for RT domain purification. RT protein was eluted with elution buffer with the presence of 50, 100 and 250 mM imidazole step-by step. In Fig-3a, a prominent 50kD band was detected
with Coomassie blue staining. Results of western blot also confirmed the purification results (Fig-3b).
Expression RT domain in reticulocyte lysate expression system
Recombinant HBV-RT was also expressed in reticulocyte lysate system as a positive control in activity assay. In terms of previous data, up to 500μg protein will be pro-duced per 50μl reaction (data not shown); therefore 5μl of product from reticulocyte lysate expression reaction was employed in enzyme activity assay.
Characterization of purified RT fraction activities and comparison with RT from in vitro expression system
To measure the enzymatic activities of purified RT do-main protein, DDDP activity and RDDP activity assays were performed under the reaction conditions described previously. The incorporation of radioactivity into DNA/RNA template-primers in solution was measured with liquid scintillation counter. Enzyme activities com-parison between RT from in vitro expression system (RT-IV) and purified RT from E.coli expression system (RT-50) was showed Fig-4.
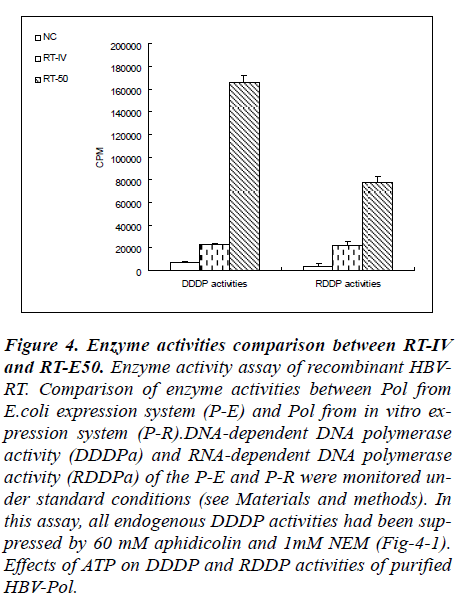
Figure 4: Enzyme activities comparison between RT-IV and RT-E50. Enzyme activity assay of recombinant HBV-RT. Comparison of enzyme activities between Pol from E.coli expression system (P-E) and Pol from in vitro ex-pression system (P-R).DNA-dependent DNA polymerase activity (DDDPa) and RNA-dependent DNA polymerase activity (RDDPa) of the P-E and P-R were monitored un-der standard conditions (see Materials and methods). In this assay, all endogenous DDDP activities had been sup-pressed by 60 mM aphidicolin and 1mM NEM (Fig-4-1). Effects of ATP on DDDP and RDDP activities of purified HBV-Pol.
Results from RT-50 showed a very high DDDP activity and a relatively low RDDP activity in these reaction con-ditions. In a contrary, results from RT-IV showed very low level both in RDDP activity and in DDDP activity. In terms of the results of enzyme activities assay, a phe-nomenon was observed that RT-50 exhibited high DNA polymerase activities no matter what the template is a DNA strand or a RNA strand. And in RT-IV group, a same phenomenon was also observed. These results indicated that DDDP activity and RDDP activity were harmonious presence in RT domain of human HBV-Pol.
Expression and purification of HBV-RT in E.coli. Ex-pression system
Expression and purification of HBV-Pol. Recombinant human HBV-RT was produced in E.coli cells transformed by pTrcHis-A-RT and then purified with nickel-based resin. A detailed explanation of the procedure was pre-sented in Materials and Methods. Protein samples ob-tained from the purification steps were analyzed by 4-12% SDS-PAGE, and then the gel was stained with Coomassie Blue (Fig. A). The HBV-RT bands were located at the ex-pected molecular mass, approximate 50kDa according to the prestain molecular weight marker. In Fig. B, although HBV-RT bands were also detected in 50mM imidazole (E50-1 and E50-2) washing fractions, mostly part of HBV-RT was eluted by buffer with 100mM (E100) and 250mM imidazole (E250) and E. Purified recombinant human HBV-Pol was also analyzed by immunoblot analy-sis with an anti-RT antibody (Fig. B). HBV-RT bands were detected in total lysate of induced transformant (TL), but not in 5th washing fraction with 10mM imidazole (W5).
Discussion
HBV polymerase plays a crucial role in HBV lifecycle. The activities are highly During HBV genome replication stage, HBV-Pol shows RDDPa and DDDPa in minus strand and plus strand synthesis, respectively. The organi-zation of HBV-Pol has been studied for many decades. Terminal protein (TP) domain triggers minus strand HBV genomic DNA in a protein-priming mechanism. RNase H domain (RH) degrades the RNA template which named progenomic RNA after minus strand of HBV genomic DNA. These two domains play important roles, but they are not the critical domain in DNA replication stage. DDDPa and RDDPa should be presence harmoniously. Therefore we expressed the RT domain of human HBV in eukaryotic expression system and prokaryotic expression system, and then compared the activities of two expressed protein products.
Expression of an active RT domain of human HBV poly-merase in heterologous systems has been carried out with limited success. Although recombinant RT had been ex-pressed in different system and purified in various ways, obtaining high purity active RT domain of HBV-Pol (HBV-RT) for biochemical study is still problematic [9,10]. It has been noted that HBV-RT was stable in het-erologous expression system and also during purification process. Inclusion body formation of HBV-RT was easily inhibited during purification step using a weak detergent.
To prevent target protein from oxidization and keep cys-teine residues (nearly 2% in total amino acid residues) in reduced condition, high concentration reducing agents were employed throughout purification steps.
In terms of results from enzyme activities, HBV-RT showed a low specific DNA polymerase activity. Molecu-lar modeling results also indicated that RT catalyzing site surrounded nucleotide template with only a few potential hydrogen bonds binding to single substrate strand [12]. Considering the function of TP domain, it has lower RDDP activity. In this study, according to the results of activity assay of HBV-RT, a higher DDDP activity was exhibited than its RDDP activity, which mainly because of RNA template-primer substrate which was partially degraded during template-primer binding steps. Human HBV-Pol is critical for HBV genome DNA replication. It showed RDDP activity in minus strand, where as DNA synthesis and DDDP activity in plus strand synthesis. Re-cent studies have proposed that some chaperones might be needed for RDDP activity at the start point of DHBV replication [15]. The RDDP activity of our DHBV P pro-tein was very weak compared with DDDP activity and the HBV P protein did not have RDDP activity. Most of ap-proved medications for the treatment of chronic hepatitis B such as lamivudine, adefovir dipivoxil, entecavir and telbivudine are targeted for HBV-RT [1]. However, the stable and large scale heterologous expression of func-tional RT domain of human HBV-Pol in common host such as E.coli or yeast has not successfully purified yet. Employing the approach in this work, a functional RT domain of human HBV-Pol was achieved in E.coli ex-pression system. The availability of this recombinant pro-tein in pure form should facilitate the antibody prepara-tion and detailed analysis of the structure and mechanism of RT domain. Large quantity of functional HBV-RT was also required in high throughout screening assay for po-tential inhibitors development.
Since the RDDP and DDDP activities of human HBV-Pol represent the critical step in the human HBV life cycle, RT domain is the most important drug target for antiviral agents. In this work, we showed the expression and purifi-cation of enzymatically active RT domain from human HBV-Pol by E.coli expression system. It is obvious that large production of functional human HBV-Pol is essen-tial, not only for antibody preparation but also for rational design of the specific inhibitors of human HBV Poly-merase.
Acknowledgements
The work was supported by the grant of Rural develop-ment administration, South Korea.
References
- Rivkin, A. Entecavir: a new nucleoside analogue for the treatment of chronic hepatitis. Drugs Today 2007; 43: 201-20.
- Shepard CW, Simard EP, Finelli L, Fiore AE, Bell BP. Hepatitis B virus infection: epidemiology and vaccina-tion. Epidemiol Rev 2006: 28: 112-125.
- Bavand M, Feitelson M. and Laub O. Hepatitis B virus infection: epidemiology and vaccination. J. Virol. 1989; 63: 1019-1021
- Lanford RE, Notvall L, Lee H, Beames B. Transcom-plementation of nucleotide priming and reverse tran-scription between independently expressed TP and RT domains of the hepatitis B virus reverse transcriptase. J Virol 1997; 71: 2996-3004.
- Wei X, Peterson DL. Expression, purification, and cha-racterization of an active RNase H domain of the hepa-titis B viral polymerase. J Biol Chem 1996; 271: 32617-32622.
- Hu J, Toft DO, Seeger C. Hepadnavirus assembly and reverse transcription require a multi-component chaper-one complex which is incorporated into nucleocapsids. EMBO J 1997; 16: 59-68.
- Wang GH, Seeger C. The reverse transcriptase of hepa-titis B virus acts as a protein primer for viral DNA syn-thesis. Cell 1992; 71: 663-670.
- Kim SS, Shin HJ, Cho YH, Rho HM. Expression of stable hepatitis B viral polymerase associated with GRP94 in E. coli. Arch Virol 2000; 145:1305-20.
- Kim Y, Jung G. Active human hepatitis B viral poly-merase expressed in rabbit reticulocyte lysate system. Virus Genes 1999; 19: 123-130.
- Choi J, Kim EE, Park YI, Han YS. Expression of the active human and duck hepatitis B virus polymerases in heterologous system of Pichia methanolica. Antiviral Res 2002; 55: 279-290.
- Sharon A, Chu CK. Understanding the molecular basis of HBV drug resistance by molecular modeling. Anti viral Res 2008; 80: 339-353.
- Das K, Xiong X, Yang H, Westland CE, Gibbs CS, Sa-rafianos SG, Arnold E. Molecular modeling and bio-chemical characterization reveal the mechanism of hepatitis B virus polymerase resistance to lamivudine (3TC) and emtricitabine (FTC). J Virol 2001; 75: 4771-4779.
- Langley DR, Walsh AW, Baldick CJ, Eggers BJ, Rose RE, Levine SM, Kapur AJ, Colonno RJ, Tenney DJ. Inhibition of Hepatitis B Virus Polymerase by Ente-cavir. J Virol 2007; 81:3992-4001.
- Jeong JH, Kwak DS, Rho HM, Jung G. The catalytic properties of human hepatitis B virus polymerase. Bio-chem Biophys Res Commun 1996; 223: 264-71.
- Beck J, Nassal M. Reconstitution of a functional duck hepatitis B virus replication initiation complex from separate reverse transcriptase domains ex-pressed in Escherichia coli. J. Virol. 2001; 75: 7410- 7419.