Review Article - Journal of Food Science and Nutrition (2021) Volume 4, Issue 9
A Review exclusively based on gut microbiome and effects of probiotic and prebiotic
Abhishek Amit*
Department of Microbiology, School of Life Sciences, Sikkim University, Samdur Sikkim, India
*Correspondence to
Dr. Abhishek Amit
Department of Microbiology, School of Life Sciences
Sikkim University
Sikkim
India
E-mail: abhishekamit62@gmail.com
Accepted on 04 October, 2021
Abstract
Many humans and non-human animals including insects, are hosts to numerous microorganisms that resides in the gastrointestinal tract as well. This gut is one niche that human microbiota inhabits. Gut microbiota of the human has the largest number of bacteria and the greatest number of species compared to other area of the body. In this review we have discussed about the human gut microbiota, diversity of microorganisms and its colonization process along with mechanisms, different signalling methods, functions, disruptions of the gut microbiota and the consequent effects. Effects of different probiotics and prebiotics have also been discussed in this review. We have found that human gut is mainly dominated by Proteobacteria, Firmicutes, Actinobacteria and Bacteroidetes. The most tried and tested manner in which the gut microbiota composition may be influenced is through the use of live microbial dietary additions, as probiotics, they also facilitate smooth functions of the intestinal environment. Most commonly used probiotic strains are: Bifidobacterium, Lactobacilli, S. boulardii and B. coagulans. Prebiotics like FOS, GOS, XOS, Inulin; fructans are the most commonly used. They are mostly fibers that are non-digestible food ingredients and beneficially affect the host’s health by selectively stimulating the growth and/or activity of some genera of microorganisms in the colon, generally Lactobacilli and Bifidobacteria.Keywords
Gut microbiota, probiotics and prebiotics.
Introduction
The word “Gut microbiota” refers to a microbial consortia which consists of trillions of microorganisms and their genetic material that reside in our intestinal tract. Microorganisms in gut, mainly comprising of bacteria, yeast, virus and archaea which are known to be involved in functions that are critical to the health and well-being [1,2]. These bacteria and other microbes inhabit in the digestive system and play a key role in digesting food and absorbing and nutrients. Other processes performed by the gut microflora include metabolism, synthesis of different essential vitamins, regulation of body weight, immune regulation as well as the brain functions and regulation of mental state along with mood.
Methods and Materials
Human gastrointestinal tract (250–400 m2) is one of the largest interfaces between the host and the microbiota and it is represented by the environmental factors and host factors/genetic susceptibility. In an average life span of human, about 60 tonnes of food passes through the human GI tract, with an abundance of microbes from the environment that impose a huge threat on gut integrity. Bacteria, eukaryote and archaea colonising on the GI tract are collectively termed as ‘gut microbiota’ and thought to be co-evolved along with the host over thousands of years forming an complex and mutually beneficial relationship[3,4]. Cumulatively gut microbiota has been estimated to exceed about 1014 in number, which encompasses ∼10 times more microbial cells than the number of human cells and over 100 times the amount of genomic content (microbiome) as compared to the human genome. A recently revised estimate has suggested that the ratio between the human cells and bacterial cells is actually closer to 1:1.
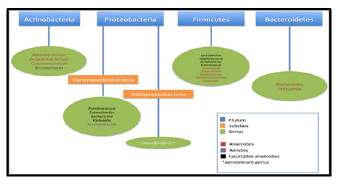
Data from the studies have identified 2172 species isolated from human gut which have been classified into 12 different phyla, of which 93.5% belongs to Proteobacteria, Firmicutes, Actinobacteria and Bacteroidetes [4,5]. Akkermansiamuciniphila sp. has been reported as the only known representative of the phyla Verrucomicrobia. In humans, 386 of the total identified species have been found as obligate anaerobes; hence these microorganisms are mostly from large intestine/colon rather than the other parts of GI tract.
The gut microbiome and particularly the study of its origin in neonates, has become subtopics of great interest within the field of genomics which includes origins and establishment of the neonatal gut microbiome and how it is affected by neonatal nutritional status (breast fed versus formula fed) and gestational age (term versus preterm).
Studies regarding animal have shown that prenatal transmission of microbes to the foetus is possible, and physiological changes among the pregnant women during the utero transfer was also observed. However, the direct evidence regarding the utero transfer of gut microbiota in humans is still lacking.
During birth, different factors influence the establishment of bacteria in our gut which begin to populate very early in life. According to some research it is suggested that this establishment begins while we are still in the womb; whether we are delivered normally or by C-section, and if we are breast or bottle-feeded that also determine the shape of the structure of our gut microbiota.
After the birth, generally the new-borns become exposed to microbes from their mother and the surrounding environment of birth process itself is the main contributor of this exposure. New-borns through vaginal delivery acquire bacteria resembling the maternal vaginal microbiome (predominantly Lactobacillus and Prevotella), whereas the new-borns from C-section acquire bacteria resembling the skin microbiome, predominantly Staphylococcus
Offspring exposed to stress, particularly during late pregnancy, shows significant reduction in the abundance of Lactobacillus and Bifidobacterium This has been observed mainly in vaginal deliveries, but not in C-section deliveries, which indicates that that delivery method has an impact on the transmission of maternal intestinal bacteria to the off-spring.
In the past few decades, the health benefits imparted by probiotics and prebiotics as well as synbiotics have been the subject of extensive research. These food supplements are termed as functional foods which have been demonstrated to alter, modify and reinstate the pre-existing intestinal flora[6]. They facilitate smooth functions of the intestinal niche. Bifidobacterium, Lactobacilli, S. boulardii, and B. coagulans are most commonly used probiotic strains. Saccharomyces, Enterococcus, Streptococcus, Pediococcus, Leuconostoc, Bacillus, Escherichia coli are some other examples of microbes which plays important role as probiotics. FOS, GOS, XOS, Inulin, fructans are the prebiotics most commonly termed as synbiotics which when used together with probiotics and are able to improve the viability of the probiotics. The technological analysis has also helped the researchers in finding out the microbial diversity from the samples, among these some are 16s rRNA sequencing, 23s rRNA sequencing, culture dependent and independent method, NGS, etc. Bioinformatics is also playing an important in computational analysis of organisms.
Human gut microbiota and its establishment
Establishment of gut microbiota: a journey from neonate to adult
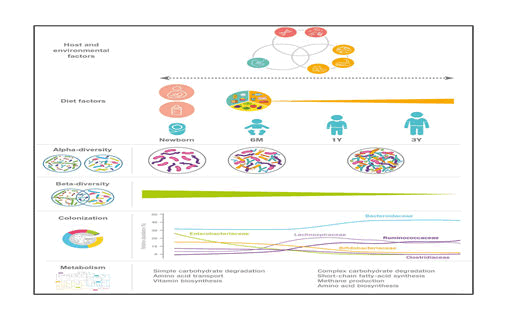
During the 1st month after birth, the microbial composition in the gut of the new-born is known to be affected by the mode of delivery, use of antibiotics, geographical location, surroundings and the type of feeding (breast/formula). Consequently, the neonatal period and early infancy has attracted much attention. The gut microbial composition continues to develop after the first period until the age of 3 years; and the 1st year has been designated as “a window of opportunity” for microbial modulation. The beginning and end of this window is currently under debate, but it likely coincides with the complementary feeding period, marking the gradual transition from milk-based infant feeding usually occurring between 6 and 24 months. Furthermore, the ‘first 1000 days,’ until the age of 2 years, are generally recognized as the period of importance for the healthy development of children.
From evidences now it is clear that the abundance of Lactobacillaceae, Bifidobacteriaceae, Enterococcaceae and Enterobacteriaceae decrease during the periods from 9 to 18 months, i.e., during the period characterized by transition from milk-based feeding to family diet while specie under the families Lachnospiraceae, Ruminococcaceae and Bacteroidaceae increase in abundance during the same period [7,8].
Breast milk keeps the gut microbiota in a state that can be characterized by a high relative abundance of Bifidobacterium along with other breast milk associated bacteria such as Veillonella, Enterococcus, Lactobacillus and Streptococcus; and these characteristics are affected only to a limited degree by mixed feeding and introduction of the first solid foods as long as the child is still partially breast-fed However, as complementary feeding progresses, the gut microbial composition changes (increase in the diverse groups of Lachnospira and Ruminococcus and decrease of Bifidobacterium, Enterobacter, Enterococcus, Lactobacillus, Veillonella, Clostridia) and total diversity increases with time
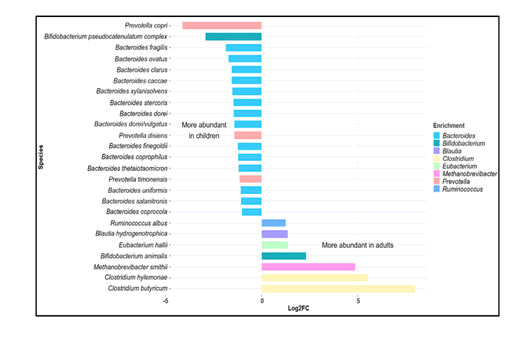
In a study on the gut microbiota of children between the age of 1 and 4 years and adults, it was found that more prevalent in children than in adults were members of the Actinobacteria, Bacilli, Clostridium cluster IV (Ruminococcaceae), and Bacteroidetes. In contrast, members of Clostridium cluster XIVa (Butyrivibrio crossotus and related bacteria) were more abundant in adults. Children within the age of 3 – 4 years have a lower microbial diversity, with a higher relative abundance of Bifidobacterium, than adults. Microbial diversity at 5 years of age is significantly lower than that in adults. At this stage taxa including Actinobacteria, Bacilli and Clostridium cluster IV retain the abundances similar to those in infants while others, such as: Clostridium cluster XIVa (Lachnospiraceae), adopting a distribution more like that in adults surveyed the gut microbiome from children within the age of 7–12 years in terms of both taxonomic and functional analyses. Bifidobacterium and Faecalibacterium spp. were found to be significantly more abundant in children than in adults, whereas adults display an enrichment in Bacteroides (i.e. Bacteroides vulgatus and Bacteroides xylanisolvens) (Figure 1).
Figure 2: Variation in gut microbiota in children and adults. Data retrieved from the study on school-age children (age 6 – 9 years) by Zhong et al. (2019) based on metagenomic shotgun sequencing at species level. The graph depicts selected major genera differing in abundance between children and adults (log2-fold change >1, < –1).
Human gut microbiota
We have entered into a revolutionary period in terms of the investigation of gastrointestinal (GI) tract microbiota. Essential groundwork has revealed their diversity and distribution patterns in the mouth, oesophagus and small intestine, with particular emphasis on the large intestine/colon
In a healthy human adult, the gut microbiota is mainly dominated by two phyla, Firmicutes. The other phyla including Actinobacteria (mainly Bifidobacterium), Proteobacteria, Verrucomicrobia and Euryarchaeota, are represented in lower concentrations. After childhood, the microbiota becomes a stable system throughout adulthood, although long-term changes resulting from diet, lifestyle gastrointestinal infections, antibiotic treatments or surgery [9].
Mouth and oral cavity
According to different habitats, including the teeth, gingival sulcus, tongue, cheeks, hard and soft palates, and tonsils, which are colonized by bacteria are present in human’s oral cavity and is a part of human oral microbiome. The microbiome of oral cavity is comprised of over 600 prevalent taxa at the species level, with distinct subsets predominating at different habitats. Microbiome of oral cavity has been extensively characterized by cultivation and culture-independent molecular methods such as 16S rRNA cloning. About 619 species level taxa in 13 phyla is included in Oral Microbiome Database of human (HOMD), which includes Actinobacteria, Bacteroidetes, Chlamydiae, Chloroflexi, Euryarchaeota, Firmicutes, Fusobacteria, Proteobacteria, Spirochaetes, SR1, Synergistetes, Tenericutes, and TM7.
Stomach
Within individuals, bacterial communities of the stomach and oesophagus showed overlapping community membership. Despite of closer proximity, the stomach antrum and corpus communities have been found to be less similar than the antrum and oesophagus [10,11]. In oesophagus, Streptococcus and Prevotella have been found to be dominant; whereas the main bacterial inhabitants of the stomach include: Streptococcus, Staphylococcus, Peptostreptococcus, Lactobacillus and different types of yeast. Helicobacter pylori – a gram negative spiral bacteria that establishes on gastric mucosa and causes chronic gastritis and peptic ulcer.
According to Migrating motor complex, Low pH, and the entero-salivary circulation of nitrate as well as external factors are several innate defences which have been shown to influence significantly the microbial composition in the stomach. Biogeographic map of the human stomach has been completed by the combination of traditional analytical tools and culture-independent molecular methods, such as temporal temperature gradient gel electrophoresis (TTGE) of polymerase chain reaction (PCR)-amplified 16S rDNA fragments and the sequencing of 16S rRNA; this has shown the wide bacterial diversity in this distinct microbial ecosystem. A series of studies have revealed the existence of a distinct stomach-associated microflora beside H. pylori.
Extremely acidic environment of the stomach theoretically prevents significant bacterial colonization; however, this does not mean that no bacteria have adapted to these extreme conditions, such as obligate or facultative acidophiles that require low pH (<4.0) to sustain [12,13]. At the phyla level, members of Firmicutes, Proteobacteria, Actinobacteria, Fusobacteria, Bacteroidetes and Gemmatimonadetes have been identified. Firmicutes and Proteobacteria are the most abundant phyla in gastric mucosa. UniFrac analysis has indicated a reduction of Actinobacteria and an increase of Campylobacter concisus (Campylobacter concisus; phylum: Proteobacteria) in stomach.
Small intestine
Small intestine contains a little number of microbes due to the proximity and influence of stomach. However alkaline condition in distal portion of small intestine supports the growth of gram-negative bacteria of Enterobacteraceae [14]. Bacterial flora in the small intestine provide the regulatory signals that enables the utility of gut; however intestinal failure may be caused due to the overgrowth of bacterial flora in small intestine.
Large intestine and colon
The large intestine contains organisms belonging over 30 identified genera and as many as 500 separate species or phenotypes. About 99 % of total bacteria in the colon are obligate anaerobes and the most abundant bacteria are members of the genus Bifidobacterium and Bacteroides; Peptostreptococcus sp., Eubacterium sp., Lactobacillus sp. and Clostridium sp. are some anaerobic gram-positive cocci, are also found in the colon and large intestine but in a lesser abundance. However, 99 % bacteria of the colon come from about 30-40 species. In healthy adults Faecalibacterium prausnitzii is found as the most common species. From the recent studies on large bowel biopsies, Bacteroides has been confirmed as the most dominant genus [15,16]. The microflora in the large intestine and colon also makes an important metabolic contribution to the synthesis of certain vitamins. Indeed, these vitamins are synthesized by several intestinal genera, including Bacteroides, Eubacterium, Propionibacterium, and Fusobacterium.
Different factors of gut microbiota
Different physiological and external factors have a profound effect on the variation of gut microbiota, which includes dietary habit, age, geographical region, pregnancy, life style and medication.
Dietary habits
Our dietary habits are based on foods, usually of plant or animal origin, that contain essential nutrients for our bodies, such as proteins, fats, carbohydrates, vitamins and minerals that can be ingested and assimilated for producing the energy required for growth and life. Diet is one of the most important factors that influence the composition and diversity of the intestinal microbiota. Information on the associations between diet and the diversity of the microbial community may vary due to researcher’s complete assessments of the intestinal microflora and strive to find more detailed taxonomic information through DNA sequencing.
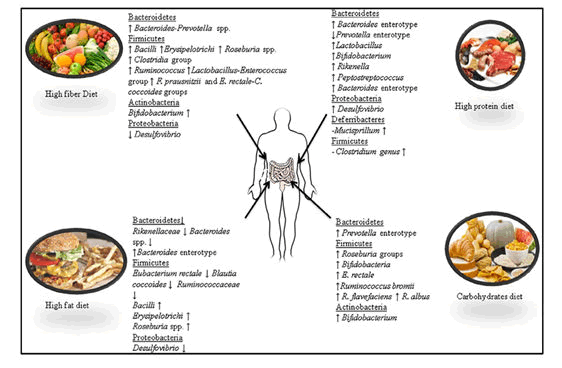
Carbohydrates are one of the major classes of biologically essential organic molecules found in all living organisms. Three types of carbohydrates: resistant starches, non-starch polysaccharides and oligosaccharides, are non-digestible and reach the gut with our daily diet. Their fermentation, specifically the fermentation of non-digestible carbohydrates, is an important activity of the gut microbiota which provides energy and drives the carbon economy of the colon. Fats are one of the three main macronutrients, along with carbohydrates and proteins, and are found in meat, poultry, nuts, milk products, butters and margarines, oils, lard, fish, grain products and salad dressings. It has been demonstrated that a high fat diet leads to a decrease in Eubacterium rectale and Blautia coccoides (Firmicutes phylum) and Bacteroides sp. from the phylum Bacteroidetes. bloom in a single uncultured clade within the class Mollicutes after assessing the consumption of a prototypic western diet (high-fat/high-sugar) that induced obesity in mice. The authors suggested that the increase in Mollicutes might reduce microbial diversity, including a reduction in the relative abundance of the genus Bacteroides. The same authors demonstrated in another study that when animals were switched from a low fat/fiber rich plant diet to a high fat/high sugar diets, they experienced a significant increase in Bacilli and Erysipelotrichi from the phylum Firmicutes and a significant decrease in members of the phylum Bacteroidetes. These results have been found to be different from those of Hildebrandt they compared wild type and resistin-like molecule beta/FIZZ2-deficient mice and assessed the influence of diet on microbiome composition. The authors showed that a high-fat diet reduced the number of gut Bacteroidetes and increased the number of Firmicutes (Clostridia group) and Proteobacteria (mainly Delta-Proteobacteria) along with the genus Desulfovibrio measured the change in caecal bacterial communities in mice which were fed a carbohydrate and high-fat (HF) diet for 12 weeks through high-throughput 16S ribosomal RNA gene sequencing. The high-fat diet caused shifts in the diversity from Firmicutes to Bacteroidetes along with the Clostridium cluster XI, XVII and XVIII.
Dietary proteins are found in foods such as meats, poultry, fish, meat substitutes, cheese, milk, nuts, legumes and vegetables. They are an important part of a balanced diet. a variety of amino acids and high animal protein intake are highly associated with the Bacteroides enterotype while the Prevotella enterotype presents the reverse association for these groups of amino acids but is associated with high values of carbohydrates and simple sugars In a study, Sprong quantified the faecal excretion of rats fed with a cheese whey protein isolate or casein, supplemented either with threonine or cysteine. The authors noted significant increases in the numbers of Lactobacillus and Bifidobacterium in faecal microbiota. In another study used high throughput DNA sequencing method and reported that mice fed with increased doses of whey protein isolate presented a significant increase in Lactobacillus, Bifidobacterium, Rikenella, Peptostreptococcus, Desulfovibrio and Mucispirillum but a significant decrease in Clostridium. The authors reported that the effects of whey protein isolates in the composition of the gut microbiota of mice were dose-dependent.
Gut microbiota of vegans and omnivores using qPCR analysis. They showed that Firmicutes (58.6% vs. 56%) and Bacteroidetes (39.1% vs. 39%) were the most abundant phyla in the vegan and omnivore group, respectively, while the phyla Verrucomicrobia, Proteobacteria, Actinobacteria, and Euryarchaeota were found to be accounted for minor proportions in both groups. However, significant differences between vegan and omnivore groups were observed with Proteobacteria being higher in the omnivore group and Verrucomicrobia being higher in the vegan group.
3.2 Geographical region
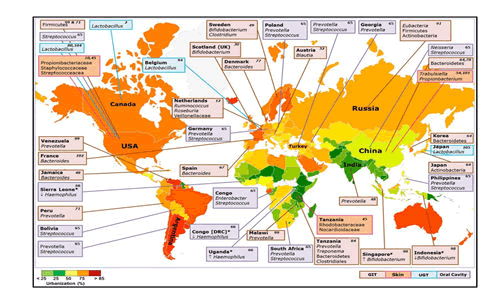
Variability in dietary behaviour is an understandable explanation for the strong influence of geographical variation on the composition of gut microbial populations. It has been demonstrated that the Bacteroides enterotype is predominant in the gut flora of people living in Western countries and consume western diet with high fat and protein content on a daily basis, while Prevotella enterotype is common in non-Western countries where the population consumes lots of fibre Microbiota appears to be similar in people living within the same area and are in contact with one another However, within the same country, geographical and socio-economical differences between localities may contribute to shaping the human gut microbiota. People living in African countries have higher gut microbiota diversity dominated by Actinobacteria (Bifidobacterium); Bacteroidetes (Bacteroides, Prevotella); Firmicutes (C. histolyticum; Eubacterium, Oscillibacter, Butyricicoccus, Sporobacter); Proteobacteria (Succinivibrio, Shigella and Escherichia) and Spirochaetes (Treponema). Meanwhile, people living in Western countries such as in Europe and America, their gut microbiota is enriched in Firmicutes (Blautia, Dorea, Roseburia, Faecalibacterium, Ruminococcus, Oscillospira, C. perfringens, C. diffusible and S. aureus), Actinobacteria (B. adolescentis and B. catenulatum), Verrucomicrobia (A. muciniphila) and Bacteroidetes (Bacteroides). Interestingly, Asian people appear to have an intermediate gut microbiota diversity, with dominant bacterial species such as Bacteroidetes (Bacteroides), Firmicutes (Prevotella) encountered in some African people and in some which are particularly dominant in Actinobacteria (B. adolescentis), Firmicutes (Butyrivibrio, Clostridium perfringens and S. aureus), which were encountered in a large proportion in Western countries. From a study on the populations in Papua New Guinea, the results of principal coordinates analysis revealed two groups, the first including Prevotella, Clostridia, Atopobium, Enterobacter, Enterococcus and Staphylococcus, and the second Bacteroides fragilis, Bifidobacterium and Lactobacillus. Differences were observed between highland and lowland participants with the former having higher numbers of most bacterial groups.
Dehinjia studied the effects of ethnicity and geography on the gut bacterial profile of Proto-Australoid tribes spread across four geographical locations (Assam, Telangana, Sikkim and Manipur) of India with distinct cultures, traditions and dietary habits. The NGS- based analysis showed that the gut microbiota of tribes in Manipur had a significantly lower Firmicutes to Bacteroidetes ratio in comparison to the tribes of Telangana and Assam. On the other hand, the phylum Actinobacteria was significantly higher in the tribes from Sikkim compared to the other tribes. The proportion of Bacteroides, Bifidobacterium, Clostridium, Enterobacter, Escherichia, Gordonibacter, Klebsiella Odoribacter, Pantoea, Parabacteroides and Slackia, representing the core gut bacterial genera, varied significantly across the Indian tribes. For example, Enterobacter, Klebsiella and Pantoea were significantly lower in the Sikkim tribes in comparison to the tribes from Assam, Telangana and Manipur, while Escherichia was more abundant in the Assam tribes than in those from Telangana and Sikkim (Dehinjia et al 2015). Ethnicity and socio-cultural practices could have an effect on the modulation of gut microbiota within inhabitants of the same geographical area.
Figure 4: The impact of non-dietary lifestyle factors on the gut microbiota has been largely ignored. Smoking and lack of exercise can significantly impact the large bowel (and potentially the microbiota) as they are risk factors for CRC (Huxley et al 2009). Indeed, smoking has a significant influence on gut microbiota composition, increasing Bacteroides-Prevotella in individuals with Crohn’s Disease (CD) and healthy individuals (Benjamin et al 2012). Smoking-induced changes in microbial populations could potentially contribute to increased risk of CD. Air-borne toxic particles can reach the large bowel via mucociliary clearance from the lungs, and increased environmental pollution associated with industrialization could contribute to concomitant increases in IBD cases (Beamish et al 2011).
Lifestyle
Another factor, lifestyle and stress, have an impact on colonic motor activity via the gut-brain axis which can alter gut microbiota profiles, including lower numbers of potentially beneficial Lactobacillus. In humans and animal models with obesity, shift in gut microbial populations has been detected, with increases in the Firmicutes and decreases in the Bacteroidetes, which could potentially contribute to adiposity through greater energy harvest. However, other data suggests the shifts in microbial populations are driven primarily by the high fat obesogenic diets. Irrespective of the cause, there are associated increases in gut bacteria linked with poor health outcomes (e.g., Staphylococcus, E. coli, and Enterobacter).
Patients with chronic alcohol overconsumption showed variability in the intestinal bacterial composition compared to those with no or very low history of alcohol intake and this was analysed by Traae Bjorkhaug and colleagues. The patients with chronic alcohol overconsumption showed more inflammatory active microbiota along with an over-representation of Proteobacteria (Gram-negative bacteria with increased concentration of endotoxins in the blood) in the gut at the phylum level and a particular increase in the genera of Clostridium, Holdemania and Sutterella (Proteobacteria). On the contrary, reported the higher abundance of the genus Faecalibacterium based on the preclinical studies among the volunteers with no or very low history of alcohol intake and Faecalibacterium is generally considered to be protective against gastrointestinal conditions.
Age
Age factor has a profound role in determining the structure of gut microbiota. A number of studies have been conducted till now to elucidate the effect of age and other age-related factors on gut community. Various age groups exhibit a different pattern of gut community structure, such as: Bifidobacterium along with Veillonella, Enterococcus, Lactobacillus and Streptococcus are dominantly found in the gut of breast-feeding neonates while Lachnospiraceae, Ruminococcaceae and Bacteroidaceae – these three-family increase in abundance during the same period. Actinobacteria, Bacilli, Clostridium cluster IV (Ruminococcaceae), and Bacteroidetes are found in children and adults but Clostridium cluster XIVa (Butyrivibrio crossotus and related bacteria) are more abundant in adults.
Functions of Gut microbiome
The importance of the gut microbiota in the development of both the intestinal mucosal and systemic immune systems can be readily appreciated from studies of Germ free (GF) (microbiota lacking) animals. Germ free animals contain abnormal numbers of several immune cell types and immune cell products, as well as have deficits in local and systemic lymphoid structures. Spleens and lymph nodes of GF mice are poorly formed. GF mice also have hypoplastic Peyer's patches (PP) and a decreased number of mature isolated lymphoid follicles. The number of their IgA-producing plasma cells have been found to be reduced, as are the levels of secreted immunoglobulins (both IgA and IgG). They also exhibit irregularities in cytokine levels and profiles and are impaired in the generation of oral tolerance.
The central role of gut microbiota in the development of mucosal immunity cannot be overlooked; considering the fact that intestinal mucosa represents the largest surface area that encounters antigens of the external environment first and that the dense populations of the gut microbiota overlying the mucosa normally accounts for the largest proportion of the antigens presented to the resident immune cells which stimulate the pattern recognition receptors [such as the TLRs and NOD-like receptors (NLRs)] of the intestinal epithelial cells. A detailed overview of the intestinal mucosal immunity can be found elsewhere. Briefly, it is composed of the gut-associated lymphoid tissue (GALT), such as the PP and small intestinal lymphoid tissue (SILT) in the small intestine, lymphoid aggregates in the large intestine and diffusely spread immune cells in the lamina propria of the GIT. These immune cells are in contact with the rest of the immune system via local mesenteric lymph nodes (MLN). In addition to the immune cells, the intestinal epithelium also plays a role in the generation of immune responses through sampling of foreign antigens via TLRs and NLRs. The mucosal immune system needs to fulfil two, sometimes seemingly conflicting, functions. It needs to be tolerant of the overlying microbiota to prevent the induction of an excessive and detrimental systemic immune response, yet it needs to be able to control the gut microbiota to prevent its overgrowth and translocation to systemic sites. Gut microbiota is intricately involved in achieving these objectives of the GIT mucosal immune system.
Mucosal/systemic immunity maturation and development
A major immune deficiency in GF animals is due to the lack of expansion of CD4+ T-cell populations. This can be completely reversed by treatment of GF mice with PSA of Bacteroides fragilis. An elegant series of experiments, showed that either mono-association of GF mice with B. fragilis or oral treatment with its capsular antigen PSA induces proliferation of CD4+ T cells. Recognition of PSA by dendritic cells (DCs) with subsequent presentation to immature T lymphocytes in MLNs was found to be required to promote the expansion. GF animals have been found to exhibit skewing towards a Th2 cytokine profile, a phenotype that was shown to be reversed by PSA treatment through the interleukin (IL)-12/Stat4 pathway. Thus exposure to a single structural component of gut microbiota promotes host immune maturation both locally and systemically. A complex microbial community containing a significant proportion of bacteria from the Bacteroidetes phylum was shown to be required for the differentiation of inflammatory Th17 cells.
Tolerance at the GIT mucosa
The GIT coexists with the dense carpet of bacteria overlying it without an induction of excessive detrimental immune activation both locally and systemically. Prevention of excessive immune response from the gut microbiota can be achieved either through physical separation of bacteria and host cells or the modifications of antigenic moieties of the microbiota to render them less immunogenic response towards tolerance.
Resident immune cells of the GIT often have a phenotypic difference from cells of the same lineage found systemically. For instance, DCs found in the intestinal mucosa preferentially induce differentiation of resident T cells into Th2 subsets, consequently promoting a more tolerogenic state in the GIT. In a series of experiments, DCs were found to be conditioned towards the tolerogenic phenotype by the stimulation of intestinal epithelial cells (IEC) through different microbial isolates, such as different Lactobacillus sp. and Escherichia coli strains. The conditioning mainly depends on the microbiota-induced secretion of TSLP and TGF-β by IEC. Interestingly, the Gram-positive Lactobacilli were more effective than the Gram-negative E. coli in conditioning the DCs towards a tolerogenic phenotype due to the greater abundance of Lactobacilli at the intestinal mucosa. Another effective mechanism of preventing colitogenic responses has been found in Bifidobacterium, which prevents the activation of the pro-inflammatory transcription factor NFκB by promoting nuclear export of a transcriptionally active NFκB subunit RelA in a PPARγ-dependent fashion. An alternate mechanism of preventing NFκB activation in response to the gut microbiota is through TLR compartmentalization have shown that while activation of basolaterally located TLR9 promotes NFκB activation, signalling originating from the apical surfaces (i.e., induced by normal gut microbiota) effectively prevents NFκB activation, promoting tolerance to the resident bacteria
In addition to microbiota-mediated tolerogenic skewing of localized immune responses, the host can also decrease the pro-inflammatory potential of microbiota constituents. The presence of the gut microbiota exposes the host to a vast amount of LPS found on the outer membranes of Gram-negative bacteria. Systemic reactions to LPS lead to highly lethal septic shock a very undesirable outcome of host-microbiota interactions. One way to avoid this disastrous scenario is to minimize the toxic potential of LPS, which is possible through dephosphorylation of the LPS endotoxin component by the action of alkaline phosphatases, specifically the intestinal alkaline phosphatase (IAP).
In addition to detoxification of LPS by IAP, IECs also acquire tolerance to endotoxin through down regulation of IRAK-1, which is essential for endotoxin signaling through TLR4 This tolerance is acquired at birth, but only in case of vaginal delivery due to the encounter with exogenous LPS during passage through the birth canal that again highlights the active role of the microbiota in tolerogenic conditioning of mucosal immune responses at the GIT.
Control of the gut microbiota
While healthy gut microbiota is essential to promote host health and well-being, overgrowth of the bacterial population may result in a variety of detrimental conditions, and different strategies are employed by the host to prevent this outcome.
Plasma cells residing at the intestinal mucosa produce secretory IgA (sIgA) that coats the gut microbiota and allows local control over their numbers. The presence of the gut microbiota is a prerequisite to activate gut DCs to induce maximal levels of IgA production as LPS augmented IgA production is negligible, comparable to the former. Furthermore, Bacteroides (Gm − bacteria) were found to be more efficient in induction of sIgA than Lactobacilli (Gm + bacteria). Interestingly, although Gm − bacteria or their structural components were also found to stimulate IgA production however the absence of intestinal IgA was found to contribute in the overgrowth of SFB, a group of Gm+ bacteria, suggesting that induction of sIgA might also be a form of competition between different members of microbiota.
Two secretory IgA (sIgA) subclasses exist: sIgA1 (produced systemically and at mucosal surfaces) and sIgA2 (produced at mucosal surfaces). sIgA2 is more resistant to degradation by bacterial proteases than sIgA1, so it is not surprising that it was found as the main IgA subclass produced in the intestinal lamina propria. Production of a proliferation-inducing ligand (APRIL) by IECs is activated via TLR-mediated sensing of bacteria and bacterial products are essential to induce switching from sIgA1 to sIgA2 production. Both Gm + and Gm − bacteria, as well as bacterial LPS and flagellin have been found as similarly effective in inducing APRIL production Thus, exposure of the gut mucosa to its neighbouring cells not only promotes IgA secretion, but also ensures that the optimally stable IgA subclass is produced. It is also of interest to note that sIgA fulfils a dual function at the intestinal mucosa, in addition to prevent the overgrowth of the gut microbiota, it also minimizes its interactions with the mucosal immune system, diminishing the host's reaction to its resident microbes.
Gut microbiota provides its host with a physical barrier to incoming pathogens by competitive exclusion, such as competition and occupation of attachment sites, consumption of nutrient bio-availability, and production of antimicrobial substances. It also stimulates the host to produce various antimicrobial compounds. Numerous AMPs, such as defensins, cathelicidins, and C-type lectins, are produced in the mammalian GIT; they are a diverse group of compounds that act by disrupting the surface structures of both commensal and pathogenic bacteria While one of the main functions of AMPs is the regulation of composition and numbers of the intestinal microbiota the interactions of AMPs and microbiota are bidirectional, as various microbial species, as well as products of microbial metabolism, have been shown to stimulate production of different types of AMPs.
Paneth cells of small intestinal crypts, express a variety of AMPs. This expression is directed by the presence of normal gut microbiota. Interestingly, while the presence of the whole microbial community was necessary to promote full levels of AMP expression, somewhat lower levels of transcripts could be induced by the presence of single bacterial species, such as B. thetaiotaomicron and L. innocua or stimulation with LPS. For induction to occur, the commensal bacteria have to be in close contact with the intestinal epithelium, as single microbial species has been found to able to produce a much higher induction when administered to RAG1/ mice (lacking secretory IgA that sequesters luminal bacteria) than to wild-type (WT) mice. The induction was mediated through TLR-MyD88 signalling.
In addition to microbial structural components, microbial metabolites also have the ability to induce AMP expression and in several cell lines. Short-chain fatty acids (SCFAs) and lithocholic acid were shown to induce the expression of cathelicidin. The induction involved the MEK/ERK pathway, AP-1 transcription factor, and histone acetylation.
Some AMPs (e.g., defensins) are initially produced in an inactive form (e.g., prodefensins), which needs to be proteolytically cleaved to be activated. Paneth cells produce matrilysin, a matrix metalloproteinase that activates defensins, and Bacteroides thetaiotaomicron colonization in GF mice was shown to induce matrilysin expression, demonstrating another aspect of microbiota-mediated induction of antimicrobial host defences.
Thus, it appears that the presence of commensal bacteria or their structural components, as well as the presence of products of bacterial metabolism have the capacity to induce the expression of AMPs and promote their activation, contributing to host protection against invading pathogens and preventing the overgrowth of the commensals themselves. Induction can be mediated through different signalling pathways, reflecting the different nature of the inductive stimuli.
The physical presence of the microbiota in the GIT also serves as a deterrent to pathogen colonization. A lot of studies, especially in the probiotics field of research, have contributed to the identification of different bacterial species with antagonistic activities against different pathogens, although the description of the exact mechanisms underlying this antagonism is often lacking.
Anaerobic faecal isolates were shown to have a greater inhibitory effect on the growth of enteric pathogens than Gm anaerobic isolates. However, the antagonistic activity was quite variable between isolates from different volunteers, as well as from different time points, highlighting the inter-individual variations of gut microbiota and its propensity for dynamic fluctuations over time.
A number of commonly utilized probiotic strains have been found to the prevent attachment and invasion of various bacterial pathogens. The Gm + Lactobacillus and Bifidobacterium, were shown to prevent Listeria infection of cultured epithelial cells through both the elaboration of secreted compounds and modulation of the epithelial cell’s immune response to Listeria. Compounds secreted by Lactobacillus were also shown to decrease colonization by pathogenic E. coli. Additionally, the presence of SFB on glial mucosa was suggested to physically exclude S. enteritidis from its attachment sites as well as to prevent colonization of enteropathogenic E. coli in rabbit.
Members of the Lactobacillus genus produce lactic acid which, in addition provides an inhibitory environment to the growth of many bacteria and potentiates the antimicrobial activity of host lysozyme by disrupting the bacterial outer membrane. Other microbial isolates from gut also produce antimicrobial substances their production is dependent on host factors which clearly indicates that the adaptation of gut microbiota to its environment. However, some microbiota isolates, specifically different species of Lactobacillus, produce antimicrobial substances that are active against a wide range of enteropathogenic bacteria, both Gm + and Gm -
Recent research has revealed several aspects of the host-microbiota interactions that promote functional and structural maturation of the GIT. To reach maturity, the GIT needs to develop efficient peristaltic motility, as well as a sufficient surface area and blood supply for nutrient acquisition. It should contain adequate attachment sites that can support the resident bacterial community, while being resistant to systemic translocation of food and microbiota-derived foreign antigens. Ultimately, the GIT needs to be able to maintain its homeostasis and regeneration following an injury.
Peristalsis and surface maturation
While previously discussed evidence from GF animals implicates the gut flora in postnatal growth of intestinal surface area, the identities of microbes/microbial molecules responsible for this development, as well as the signalling pathways through which it is stimulated, remain elusive. A number of microbiota members have been shown to induce transcription of angiogenin-3, a protein with angiogenic activity. It is intriguing that while colonization of ex-GF mice with Bacteroides thetaiotaomicron (a prominent member of post weaning gut microbiota) resulted in the same transcriptional levels of angiogenin-3 as those observed in SPF mice, colonization with Bifidobacterium infantis (a pioneer of the GIT commonly found in new-born microbiota) resulted in lower transcription of angiogenin-3. This finding suggests that temporal maturation of the gut microbiota is at least partially responsible for sequential maturation of the GIT. Bacteroides thetaiotaomicron induced angiogenesis was shown to depend on signalling via Paneth cells. Colonization of ex-GF mice with Bacteroides thetaiotaomicron was also shown to influence transcription of various host factors involved in function of the enteric nervous system, suggesting that it can modulate postnatal development of peristalsis.
Carbohydrate moieties frequently serve as microbial attachment sites or nutrient sources, making mucosal glycosylation patterns an important factor in colonization of GIT by the gut microbiota. While the host has innate mechanisms to regulate the spatial and cell-specific distribution of glycan expression, indicating that the host is armed with glycosyl transferases necessary for glycosylation processes, the glycosylation patterns are further modified by the presence of the gut microbiota. Microbiota-induced modifications happen at both the cellular (quantitative and qualitative differences in surface glycan expression on different cell types) and the subcellular (modifications of trafficking of glycan-bearing structures) levels, and it has been shown that B. thetaiotaomicron secretes a signalling molecule that induces the host to express fucose on cell surface glycol-conjugates, which can then be released and consumed by Bacteroides thetaiotaomicron. This finding demonstrates that the gut microbiota is able to generate a suitable physiological niche by modulating the intestinal glycocalyx structure.
Barrier fortifications and regenerative capacity
Preservation of homeostasis at the intestinal mucosa should be in the gut microbiota’s best interest, as it provides a convenient long-term habitat. It should not be surprising then that various microbiota members contribute to the maintenance of intestinal epithelium barrier integrity through maintenance of cell-to-cell junctions and promotion of epithelial repair following injury.
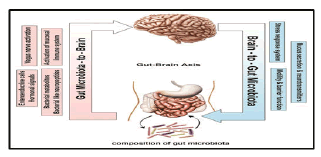
Bacteroides thetaiotaomicron has been shown to induce expression of sprr2a, important in desmosome maintenance. The expression was increased at the epithelial villus, suggesting its role in barrier maintenance. Several probiotic strains of Lactobacillus have been proved as a potential contributor to the maintenance of tight junctions in intestinal epithelia, providing protective effect in the face of pathogen assault or intestinal injury. Furthermore, signalling via TLR2, which is principally stimulated by microbial cell wall peptidoglycan, was shown to promote the integrity of the intestinal epithelium through maintenance of tight junctions and decreased apoptosis. Microbiota signalling through mucosal TLRs was also shown to be required for maintenance of intestinal epithelial homeostasis and repair following intestinal injury. Over the years, the ability of microbiota to impact on brain function has been subject to an intense research following the observation that administration of oral antibiotics and laxatives resulted in a dramatic improvement in patients with hepatic encephalopathy The observation that common categories of GI diseases (functional and inflammatory) often display correlation with psychiatric comorbidity that includes depression and anxiety in up to 80% of patients, supports the possibility that alteration of microbiota can affect CNS function. These clinical findings are supported by results from animal studies showing that certain pathogenic enteric bacteria, during the initial phase of infection, can induce an anxiety like behaviour. Furthermore, microbiota has also been shown to modulate the levels of adrenocorticotrophic hormone (ACTH) in mice. The findings have been recently corroborated by and in which germ free (GF) mice displayed increased motor activity, reduced anxiety and decreased N-methyl-d-aspartate receptor subunit expression. Modification of microbiota by a combination of probiotics has been shown to reduce anxiety in animals and bring beneficial psychological effects with a decrease in serum cortisol in patients. Furthermore, showed that long lasting treatment of mice with the probiotic bacterium Lactobacillus rhamnosus appears to impact on emotional behaviour and the expression of the neurotransmitter GABA (γ-aminobutyric acid) in the CNS in a region-dependent manner. Likewise, Lactobacillus rhamnosus treatment may be associated with a decrease in corticosterone levels as well as anxiety- and stress-related behavior. These alterations were not observed in vagotomised mice, indicating the vagus nerve may be the part of the communication pathways that allow communication between the gut and the brain. By analogy, (2009) have observed a temporal association between diet-induced alterations in intestinal microbiota diversity and changes in working and reference memories.
Figure 5: Bidirectional communication between the gut microbiota and the central nervous system (CNS). The composition of gut microbiota could modulate the function of CNS through various communication means including neural (vagus nerve activation), hormonal (enteroendocrine cells and bacterial neuropeptides), humeral (bacterial metabolites) and immunological (activation of mucosal immune system). The brain-to-gut microbiota axis is mediated via stress factors, alteration in intestinal permeability and motility and through release of neurotransmitters and mucus.
The gut microbiota mainly derives their nutrients from dietary carbohydrates. Fermentation of the carbohydrates through proximal digestion that generate indigestible oligosaccharides by colonic organisms such as Bacteroides, Roseburia, Bifidobacterium, Fecalibacterium, and Enterobacteria, result in the synthesis of short chain fatty acids (SCFA) such as butyrate, propionate and acetate, which are rich sources of energy for the host. This host energy balance is believed to be mediated via a ligand-receptor interaction of the SCFAs with a G-protein coupled receptor GPCR 41. Another entero-endocrine hormone PYY (Peptide Tyrosine Tyrosine/Pancreatic Peptide YY3-36) has also been found to implicate this action. Furthermore, butyrate can prevent the accumulation of toxic metabolic byproducts such as D-lactate. Members of the genus Bacteroides, which are the largest contributor to carbohydrate metabolism, perform this by expressing enzymes such as glycosyl transferases, glycoside hydrolases and polysaccharide lyases. The best example among these organisms is Bacteroides thetaiotaomicron, which is endowed with a genome that codes for over 260 hydrolases, which is far more than the number encoded by the human genome. The oxalate that is synthesized in the intestine as a result of carbohydrate fermentation and bacterial metabolism is countered by organisms such as Oxalobacter formigenes, Lactobacillus sp., and Bifidobacterium sp. thereby reducing the risk of formation of oxalate stone in the kidney.
The gut microbiota has also been shown to impart a positive impact on lipid metabolism by suppressing the inhibition of lipoprotein lipase activity in adipocytes. Furthermore, Bacteroides thetaiotaomicron has been demonstrated to augment the efficiency of lipid hydrolysis by up regulating expression of a colipase that is required by pancreatic lipase for lipid digestion. The gut microbiota is also enriched with an efficient machinery that helps in protein metabolizing function via the microbial proteinases and peptidases in tandem with human proteinases. Several amino acid transporters on the bacterial cell wall facilitate the entry of amino acid from the intestinal lumen into the microbiota, wherein several gene products convert the amino acids into small signalling molecules and antimicrobial peptides (bacteriocins). Important examples include conversion of Lhistidine to histamine by the bacterial enzyme histamine decarboxylase, which is coded by the bacterial hdcA genes and glutamate to gamino butyric acid (GABA) by glutamate decarboxylases, which are coded by the bacterial gadB genes. Synthesis of vitamin K and components of vitamin B are another major metabolic function imparted by gut microbiota. Members of genus Bacteroides have been shown to synthesize conjugated linoleic acid (CLA) that is known to be anti-diabetic, anti-atherogenic, anti-obesogenic, hypolipidemic and have immunomodulatory properties. The gut microbiota, especially Bacteroides intestinalis and Bacteroides fragilis have been reported to deconjugate and dehydrate the primary bile acids and convert them into the secondary bile acids and lithocolic acids in the human colon. The normal gut microbiota has also been shown to impart a healthy metabolome by increasing the concentrations of various organic acids, all of which are indicators of higher energy metabolism.
Metabolism and degradation of xenobiotics by gut community have been reported earlier. Recent studies have shown that a gut microbial metabolite pcresol can reduce the capacity of the liver to metabolize acetaminophen due to competitive inhibition of hepatic sulfotransferases. Furthermore, cardiac glycosides like digoxin have been recently shown to upregulate a cytochrome containing operon in the common organism Eggerthella lenta from the phylum Actinobacteria, which results in inactivation of digoxin. Another interesting example of microbiome induced drug metabolism is the microbial βglucoronidase induced deconjugation of the anticancer drug irinotecan that can contribute to its toxicities such as diarrhea, inflammation and anorexia .
Probiotics
Bacteria and/or yeasts are a part of probiotic supplements, foods, and beverages contain. Up until the 1960s, the only gut microflora that they were able to identify were clostridia, lactobacilli, enterococci, and E. coli. Since then, innovative techniques have helped us to discovered many more bacteria.
For potential probiotic strains screening many tests are performed. Determination of the taxonomic classification, which may give an indication of the origin, habitat and physiology are the first step for selection of a probiotic LAB strain. All these characteristics have important consequences on the selection of the novel strains (Morelli 2007). The specificity of probiotic action is more important than the source of microorganism and this was suggested by an FAO/WHO (2002) expert panel. This conclusion was given forward due to uncertainty of the origin of the human intestinal microflora since the infants are borne with virtually sterile intestine. It was also underlined by the panel that there is a need for improvement of tests to predict the performance of probiotics in humans. An ideal probiotic strain remains to be identified for any given indication though many probiotics meet criteria such as acid and bile resistance and survival during gastrointestinal transit. Furthermore, it seems unlikely that a single probiotic will be equally suited to all indications; selection of strains for disease-specific indications will be required (Shanahan 2003).
Phenotype and genotype stability, including plasmid stability; carbohydrate and protein utilization patterns; acid and bile tolerance and survival and growth; intestinal epithelial adhesion properties; production of antimicrobial substances; antibiotic resistance patterns; ability to inhibit known pathogens, spoilage organisms, or both; and immunogenicity are some next important criteria for the screening and selection of probiotics. The ability to stick to the intestinal mucosa is one among the more important selection criteria for probiotics because adhesion to the intestinal mucosa is taken into account to be a prerequisite for colonization.
The human alimentary canal is inhabited by a posh and dynamic population of around 500-1000 of various microbial species which remain during a complex equilibrium. It has been estimated that bacteria account for 35–50% of the quantity content of the human colon. Bacteroides, Lactobacillus, Clostridium, Fusobacterium, Bifidobacterium, Eubacterium, Peptococcus, Peptostreptococcus, Escherichia and Veillonella are some of them which is part of these colon. Bifidobacterium and Lactobacillus species are two bacterial strains with identified beneficial properties include mainly. The dominant microbial composition of the intestine has been shown to be stable over time during adulthood, and the microbial patterns are unique for each individual. However, there are numerous external factors that have potential to influence the microbial composition within the gut as host genetics, birth delivery mode, diet, age, antibiotic treatments and also, other microorganisms as probiotics. There are several different kinds of probiotics, and their health benefits are determined by the job that they do in the gut. Here may be a list of probiotics and their possible health benefits.
A. Lactobacillus
There are more than 50 species of Lactobacilli. These Lactobacillus is naturally found in the digestive, urinary, and genital systems. Foods like yogurt which is a fermented product, and dietary supplements also contain these bacteria. This has been used for treating and preventing a wide variety of diseases and conditions.
Some of the them are found in foods and supplements are Lactobacillus acidophilus, L. acidophilus DDS-1, Lactobacillus bulgaricus, Lactobacillus rhamnosus GG, Lactobacillus plantarium, Lactobacillus reuteri, Lactobacillus salivarius, Lactobacillus casei, Lactobacillus johnsonii, and Lactobacillus gasseri and many more.
Studies have shown some benefits linked to Lactobacillus and treating and/or preventing yeast infections, bacterial vaginosis, urinary tract infection, irritable bowel syndrome, antibiotic-related diarrhea, traveller’s diarrhea, diarrhea resulting from Clostridium difficile, treating lactose intolerance, skin disorders (fever blisters, eczema, acne, and canker sores), and prevention of respiratory infections.
B. Bifidobacterium
There are approximately 30 species of Bifidobacteria. They make up most of the healthy bacteria in the colon. They appear in the intestinal tract from the day of birth, especially in breastfed infants and are thought to be the best marker of intestinal health.
Some of this is used as probiotics which are Bifidobacterium bifidum, Bifidobacterium lactis, Bifidobacterium longum, Bifidobacterium breve, Bifidobacterium infantis, Bifidobacterium thermophilum and Bifidobacterium pseudolongum.
Studies have also shown that it can help with improving blood lipids and glucose tolerance. These have been shown to effectively alleviate IBS and significantly improve IBS symptoms like pain/discomfort, distension/bloating, urgency, and digestive disorders.
C. Saccharomyces boulardii
Saccharomyces boulardii is the only yeast probiotic. Some studies have shown that it is effective in preventing and treating diarrhea associated with the use of antibiotics and traveller’s diarrhea. It has also been reported to prevent the reoccurrence of C. difficile, treating of acne, and reducing the side effects of treatment for H. pylori.
D. Streptococcus thermophilus
This produces large quantities of the enzyme lactase, making it effective, consistent with some reports, within the prevention of lactase deficiency.
E. Enterococcus faecium
This is normally found in the intestinal tract of humans and animals.
F. Leuconostoc species
This species has been used extensively in processing of food throughout human history, and ingestion of foods containing live bacteria, dead bacteria, and metabolites of these microorganisms has taken place for a long time.
Major actions of probiotics include enhancement of the epithelial barrier, increased adhesion to i ntestinal mucosa, and concomitant inhibition of pathogen adhesion, competitive exclusion of pathogenic microorganisms, production of anti-microorganism substances and modulation of the immune system.
The intestinal epithelium is the largest mucosal surface in the human body, provides an interface between the external environment and the host. The gut epithelium is constantly exposed to foreign microbes and antigens derived from digested foods. Thus, the gut epithelium acts as a physical barrier against microbial invaders and is equipped with various elements of the innate defense system. In the gut, two key elements govern the interplay between environmental triggers and therefore the host: intestinal permeability and intestinal mucosal defense. Resident bacteria can interact with pathogenic microorganisms and external antigens to guard the gut using various strategies.
According to the widely accepted definition of a probiotic, the probiotic microorganism should be viable at the time of ingestion to confer a health benefit. Although not explicitly stated, this definition implies that a probiotic should survive alimentary canal passage and, colonize the host epithelium. A variety of traits are believed to be relevant for surviving GI tract passage, the most important of which is tolerance both to the highly acidic conditions present in the stomach and to concentrations of bile salts found in the small intestine. These properties have consequently become important selection criteria for brand spanking new probiotic functionality. In addition to tolerating the tough physical-chemical environment of the alimentary canal, adherence to intestinal mucosal cells would be necessary for colonization and any direct interactions between the probiotic and host cells resulting in the competitive exclusion of pathogens and/or modulation of host cell responses. Moreover, as enteropathogenic Escherichia coli binds to epithelial cells via mannose receptors, some probiotic strains with similar adherence capabilities can inhibit pathogen attachment and colonization at these sites of binding and thereby protect the host against infection.
Probiotic bacteria are capable to antagonize pathogenic bacteria by reducing luminal pH, inhibiting bacterial adherence and translocation, or producing antibacterial substances and defensins. One of the mechanisms by which the gut flora resists colonization by pathogenic bacteria is by the assembly of a physiologically restrictive environment, with regard to pH, redox potential, and hydrogen sulphide production. Probiotic bacteria decrease the luminal pH, as has been demonstrated in patients with ulcerative colitis (UC) following ingestion of the probiotic preparation VSL3. In a fatal mouse Shiga toxin-producing E. coli O157:H7 infection model, the probiotic Bifidobacterium breve produced a high concentration of acetic acid, consequently lowering the luminal pH. This pH reduction was related to increased animal survival.