Research Article - Biomedical Research (2018) Volume 29, Issue 21
A five-year retrospective study of the antimicrobial susceptibility pattern of Pseudomonas aeruginosa ICU clinical isolates in Al-Ahsa, Saudi Arabia.
Lorina Ineta Badger-Emeka1*, Promise Madu Emeka2 and Sayed Quadri1
1Department of Biomedical Sciences, Microbiology Department, College of Medicine, King Faisal University, Al-Ahsa. Kingdom of Saudi Arabia
2Pharmaceutical Sciences Department, College of Clinical Pharmacy, King Faisal University, Al-Ahsa. Kingdom of Saudi Arabia
- *Corresponding Author:
- Lorina Ineta Badger-Emeka
Department of Biomedical Sciences
Microbiology Department
College of Medicine
King Faisal University
Kingdom of Saudi Arabia
Accepted date: December 14, 2018
DOI: 10.4066/biomedicalresearch.29-18-1154
Visit for more related articles at Biomedical ResearchAbstract
Pseudomonas aeruginosa an opportunistic pathogen of clinical importance has been associated with infections in intensive care units worldwide. This study seeks to evaluate its susceptibility pattern over a five-year period in line with the recommended regional monitoring. Isolates were those of obtained from patients in intensive care units. Bacteria isolation was by basic bacteriological and biochemical methods. The VITEK 2 compact automated system (BioMerieux, Marcy L’Etoile, France) was used for ID confirmation and antimicrobial susceptibility assay. Data was analysed and represented as percentages as well as mean percentage ± standard error of mean (SEM). A total of 580 samples were used for the investigation. Fifty two percent (52%) of them were from males while 48% were from females. Age distribution data shows that P. aeruginosa infection was seen to be more associated in patients within the age range of 51 y and above with a total incidence was 81.25%. Colistin maintained 100% sensitivity to this bacterium throughout the duration of the study. However, for the beta lactam agents, fluoroquinolones, aminoglycosides and tigecycline, there was a decline in sensitivity from 70.4% in 2013 to 33.1% in 2017. Consequently, there was an observed growing resistance to anti-Pseudomonal drugs, from 14.7% in 2013 to 50.8% in 2017, representing a 3.5 fold increase that is alarming. Also, the mean MAR index of the antibiotic susceptibility was 0.43. The obtained results point to possibility of the isolates originating from a high risk source of contamination region where there high antibiotics use.
Keywords
Pseudomonas aeruginosa, Antibiotic resistance, Susceptibility, Sensitivity, Infection, Isolates
Introduction
The genus Pseudomonas consists of more than 120 species present in our environment infecting plants, animals and humans [1]. Of these species, Pseudomonas aeruginosa has been associated with human infections despite the fact that it exists naturally in the environment [2,3]. Recognised as an opportunistic pathogen and a leading cause of nosocomial infections in immunocompromised patients [4,5], P. aeruginosa has become of enormous clinical importance [6]. The bacterium is associated with Intensive Care Unit patients were in some cases is the causative organism associated with UTIs in catheterized patients in which the catheters are reported to be the source of host entry [7]. Being the leading cause to both nosocomial and community infections worldwide, Pseudomonal infections have been receiving much attention by researchers in recent years: Fatimah et al. [8] for respiratory tract infections; Mansoor et al. [9] for otitis media; Kim et al. [10] for wound infections Raffaele et al. [11] for chronic wound infections, amongst a wide range of other hospital and community acquired infections. Chronic lung infections in cystic fibrosis patients [12], hospital acquired pneumonia (HAP) in immunocompromised patient [13] have all been linked to P. aeruginosa, resulting in high mortality and morbidity rates in these patients. The European Center for Disease Prevention and Control 2013 [14] report indicates that 9% of all healthcare associated infections was attributed to P. aeruginosa with more researchers [15,16] reporting on this opportunistic nosocomial pathogen.
With a rise in resistance to antimicrobials in the 21st century, Pseudomonas aeruginosa has not been exempted from this public health problem. According to Yadov et al. [17] advances in medical and surgical healthcare facilities with the introduction of antimicrobial agents having anti-Pseudomonal activities have not been able to avert or reduce life threatening infections caused by Pseudomonas aeruginosa nor their complications in hospital acquired infections [18]. From the time of Mayhall’s [19] report on the complications resulting from hospital acquired infections due to Pseudomonas aeruginosa, this Gram negative bacterium belonging to the family Pseudomonadaceae has risen to become a challenging pathogen of global importance with reports on high resistance to available antimicrobials [16,20]. Of a particular worrying trend are the isolates which have been associated with ICU infections [21]. Earlier Samporn et al. [22] stipulated that the high antimicrobial resistance associated with ICU isolates has resulted in high morbidity and mortality rates being attributed to Pseudomonal infections. The ability for P. aeruginosa to be a leading pathogen in many diverse HAI and CAI is attributed to a number of reasons one which is it’s ubiquitous nature at being able to survive in moist environments as well as being able to resist antiseptics and antimicrobial [23]. There is also the result of significant changes in the evolving microbial genetics making P. aeruginosa multifactorial with genes encoding porins, efflux pumps amongst others all of which have contributed to different mechanism of resistance to antimicrobials as reported by Ozer et al. [24]. The impermeability of this Gram negative bacterium naturally contributes to the ability to resist many antibiotics [25]. There is also the ability of the bacterium to easily acquire resistance thus creating challenging therapeutic scenarios that has led to failure of all beta-lactam antibiotics to eradicate P. aeruginosa strains [26,27].
As the world tries to tackle the problem in the rise of resistance to antimicrobials, the need for continuous regional monitoring cannot be over emphasized. The present investigations looks at a five year antibiotic susceptibility pattern exhibited by Pseudomonas aeruginosa isolated from patients in intensive care units in Al-Ahsa, South-eastern region of Saudi Arabia. And also, to assess the extent of its growing resistance to current antibiotics over same period. This is with a view of providing an antibiotic monitoring update on this bacterium in this region of the world.
Materials and Methods
Ethical consideration
No ethical approval was required as samples were part of routine laboratory diagnosis for the care of patients. There was no need for a written informed consent as all the patients were anonymous and no personal information was not used in the study.
Sample source, inclusive and exclusive criteria
The samples used for the study were collected from 2013 to 2017. They were isolates from both male and female patients who were in ICUs with all samples being part of routine laboratory diagnosis. All samples were grouped on the following patient demographic data: age group, gender and the source of sample collection. Only samples obtained from intensive care units were used for the study and were irrespective of patient’s neither age nor gender.
Microbial isolation and antibiotic susceptibility test
Collected specimen included blood, urine, endotracheal aspirates, sputum and wound. Basic bacteriological culturing method was used for bacteria isolation and microbial culturing. Blood and MacConkey agars were used for the culturing of urine samples. In addition to these two media, chocolate agar was used in the culturing of blood, sputum and tracheal aspirates [28]. All cultures were incubated aerobically at 37°C for 24 h. Pure cultures of isolates were used for the identification. Routine Biochemical methods described by Collins et al. [29] and Forbes et al. [30] were used for the identification of Pseudomonas aeruginosa. They were nonlactose fermenting, oxidase positive, Gram-negative Bacilli. The disk diffusion method was used for antimicrobial susceptibility test by plaiting of isolates on Muller Hilton agar using the M02-A11 of the Clinical Laboratory Standards Institute (CLSI) [31]. The following antibiotic disks were used for the study: amikacin (30 μg), colistin (10 μg), gentamicin (10 μg), piperacillin/tazobactam (100/10 μg), amoxicillin/clav. acid (20/10 μg), azithromycin (15 μg), cefipime-protec, cefotaxime (30 μg), ceftazidime (30 μg) ceftizoxime (30 μg), ceftriaxone (30 μg), ciprofloxacin (5 μg), imipenem (10 μg), levofloxacin (5 μg), meropenem (10 μg), minocycline (30 μg), rifampicin (5 μg), tetracycline (30 μg), nalidixic acid (30 μg), nitrofurantoin (300 μg), tigecycline (15 μg), doxycline hydrochloride (30 μg), trimeth/sulphamethoxazole (1.25/23.75 μg). Results of disc diffusion tests were interpreted using the M100-S25 of CLSI [31]. Further identification of the isolates was carried out using the VITEK 2 compact automated system (BioMerieux, Marcy L’Etoile, France) using the GN Cards according to the manufacturers’ guidelines.
Determination of multi-antibiotic resistance index
The MAR index was calculated for Pseudomonas aeruginosa isolates that showed resistance to more than three antibiotics as in the method described by [32-34]. This was considered as the number of antibiotics to which the tested isolates were resistant to, divided by the total number of antibiotics to which the organism was tested against for sensitivity [28].
Statistical analysis
Data obtained are represented as percentages and also as mean percentage ± standard error of mean (SEM). T-test was used to determine significant difference between isolates from different sources and degree of resistance using GraphPad Prism software. P-values were then calculate and statistical significance was considered at p<0.05.
Results
Demography and source of P. aeruginosa isolates
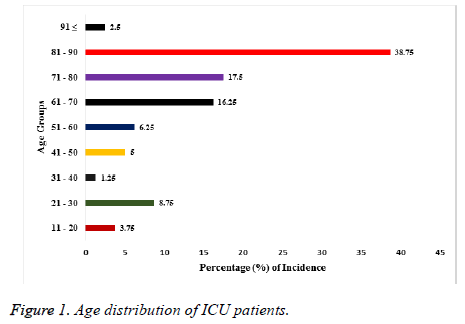
A total of 580 samples were used for the investigation. Fifty two percent (52%) of them were from males while the remaining 48% were from females. The age range of the patients from whom samples had been collected is shown in Figure 1. The youngest patients were within the age range of 11-20 y and they made up 3.75% of all the samples. The oldest patients were 91 y and above with a percentage of 2.5. Majority (38.75%) of the samples were from patients between the ages of 90 to 81 years. While those of the following age groups 80-71, 70-61, 60-51 were 17.5%, 16.25% and 6.25% respectively as shown in Figure 1. The distribution of these isolates as regards the source of specimen isolation is shown in Table 1. Forty six percent (46.5%) of the total isolates were from sputum, while those from wound, urine, blood and ENT represented 27.9%, 11.6%, 2.4% and 11.6% respectively. The table also shows the mean percentage resistance exhibited by the isolates against the tested antibiotics for the period of 2013 to 2017, calculated as mean ± SEM. Isolates from sputum were more resistant (36.5%) significantly (p<0.05†) for the 5 y period compared to samples from ENT and blood sources. While isolates from blood specimens which were the least in number, also had least mean percentage resistance for the same period under review.
| Isolate source | Total number of isolates | Percentage (%) | Mean percentage resistance (± SEM) | P-value |
|---|---|---|---|---|
| Sputum | 270 | 46.5 | 36.5± 9.77 | 0.001† |
| ENT | 67 | 11.6 | 13.8± 3.68 | 0.00218* |
| Wounds | 162 | 27.9 | 23.7± 6.33 | 0.072 |
| Urine | 67 | 11.6 | 23.7± 6.35 | 0.072 |
| Blood | 14 | 2.4 | 11.5± 3.09 | 0.0134* |
| †Comparing sputum isolates with group of other sources (p<0.05); *Comparing sputum isolates with individual sources (p<0.05) | ||||
Table 1. Distribution of P. aeruginosa isolates based on sampling source.
Antimicrobial susceptibility
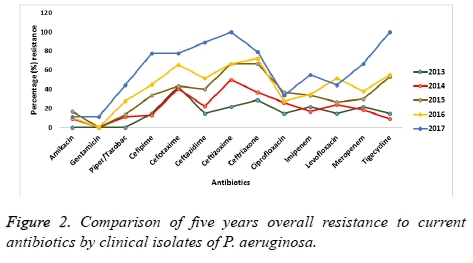
A five year observation on the antimicrobial susceptibility by P. aeruginosa against the tested antibiotics is presented in Figure 2. Colistin maintained a complete (100%) sensitivity throughout the period of observation. However, for the beta lactam agents, fluoroquinolones, aminoglycosides and tigecycline, there was a decline in sensitivity from 2013 to 2017. Resistance is seen to have increased to these antimicrobials over the five-year period of this study. Antibiotics such as amikacin and gentamicin exhibited the least resistant with about 11.1% each after 5 years. However, P. aeruginosa isolates became completely (100%) resistant to ceftazidime and tigecycline by the year 2017 while the Fluoroquinolones were seen to have had a better antimicrobial susceptibility profile against the isolates than that exhibited to the carbapenems.
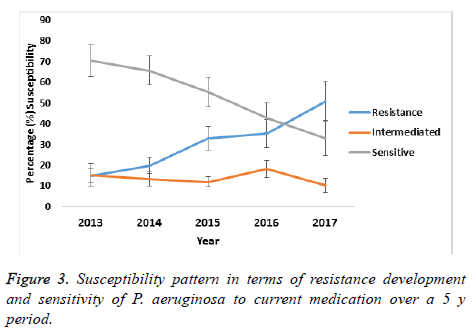
A comparison on the mean susceptibility pattern in terms of decreasing sensitivity and increasing resistance for the period of 2013 to 2017 showed that the buffer zone representing the intermediate susceptibility of P. aeruginosa did not change significant between 2013 and 2014, indicating that transition to resistance was minimal as is shown in Figure 3. However, from 2015 to 2017 this transition to resistance increased and at the same time, the sensitivity of P. aeruginosa decreased as shown in the figure. Thus pointing to an overall gradual development of resistance to antibiotics over the five years period of study.
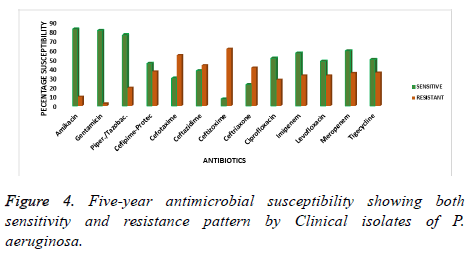
The overall mean of the five years observation is presented as the characteristic susceptibility of P. aeruginosa from 2013 to 2017. This has been represented by the mean percentage of sensitivity and resistance to current antibiotics as shown in figure 4. Amikacin, gentamicin and piperacillin/tazobactam showed a better sensitivity profile and hence less percentage resistance as compared to all the other antibiotics. The antimicrobial sensitivity pattern for these drugs was 82.3, 80.7 and 76.1% respectively over the years of study. However, the isolates were less sensitive to ceftizoxime, ceftriaxone and ceftazidime with a percentage resistance of 61, 40.9 and 43.4% respectively as is shown in Figure 4. Indicating that these antibiotics became less effective over time in the treatment of P. aeruginosa infections.
There was a noticeable variation as regards antimicrobial susceptibility and the specimen source of P. aeruginosa isolation. In Table 2, the isolates from different specimens displayed varied resistant pattern to the antibiotics used in their treatment. The table shows sputum isolates to be more resistant to the cephalosporins than to the aminoglycosides. On the other hand, isolates from blood, urine wound and ENT were more susceptible to imipenem than to the other beta lactam antibiotics against which they were tested with the exception of colistin.
| Antibiotics | Sources of isolates | ||||
|---|---|---|---|---|---|
| Blood | Urine | Sputum | Wound | ENT | |
| Amikacin | 0 | 0 | 10 | 8 | 0 |
| Colistin | 0 | 0 | 0 | 0 | 0 |
| Gentamicin | 0 | 19.4 | 0 | 8.6 | 0 |
| Piperacillin/tazobactam | 0 | 19.4 | 37.4 | 16.7 | 19.4 |
| Cefipime-protec | 0 | 29.9 | 32.6 | 12.3 | 10.4 |
| Cefotaxime | 0 | 19.4 | 52.6 | 45.7 | 40.2 |
| Ceftazidime | 100 | 40.2 | 40 | 45.7 | 40.2 |
| Ceftizoxime | 0 | 50.7 | 70 | 50 | 29.9 |
| Ceftriaxone | 0 | 40.2 | 40 | 50 | 19.4 |
| Ciprofloxacin | 0 | 19.4 | 37.4 | 12.3 | 19.4 |
| Imipenem | 0 | 0 | 40 | 16.7 | 0 |
| Levofloxacin | 0 | 10.4 | 50 | 21 | 0 |
| Meropenem | 0 | 40.2 | 37.4 | 12.3 | 0 |
| Tigecycline | 50 | 19.4 | 37.4 | 16.7 | 0 |
Table 2. Showing percentage (%) resistance variations by P. aeruginosa from different sources of isolation.
The results on the MAR index for the P. aeruginosa isolates are shown in Table 3. For majority (91%) of the isolates, the MAR index was above an index of 0.2 with a mean of 0.434.
| MAR index | Number of isolates encountered |
|---|---|
| 0 | 0 |
| 0.1 | 3 |
| 0.2 | 49 |
| 0.3 | 203 |
| 0.4 | 136 |
| 0.5 | 50 |
| 0.6 | 54 |
| 0.7 | 37 |
| 0.8 | 31 |
| 0.9 | 7 |
| 1 | 10 |
Table 3. A five year multi-antibiotic resistance index (MAR) for P. aeruginosa.
Discussion
Pseudomonas aeruginosa is listed among the Gram negative bacteria that are multidrug resistant and of major public health concern in the 21st century [35]. The ubiquitous characteristic of this opportunistic pathogen [36], its presences in healthcare settings and the complexity by which the chromosomal encoded mechanisms are regulated or co-regulated has made this bacterium one of the greatest therapeutic challenges in this era [35]. That the isolates used in the present investigation were from patients in intensive care units (ICUs) points to the possibility that they had been of nosocomial in origin due to the resulting antimicrobial susceptibility pattern exhibited by the isolates. This resultant antimicrobial susceptibility pattern could be attributed to a number of varying reasons. Yusuf et al. [16] reported on the emergence of MDR P. aeruginosa isolates in ICU patients and this they stipulated was based on the duration of antibiotic exposure in the ICUs. Although all the isolates in the present investigation were form patients in the ICUs, the length of patient’s exposure to these antibiotics was not ascertained. A wide range of anti-Pseudomonal drugs is seen as having been been used in the treatment of these ICU infections. The obtained results on the susceptibility of the isolates against antibiotics such as the β-lactams, fluoroquinolones, aminoglycosides and tigecycline showed a decline in the sensitivity from 2013 to 2017 as shown in Figure 2. This probably points to the fact that although the bacterium had been sensitive to the antibiotics at the start of the research, there had been a gradual drift over the years towards resistance, further highlighting to need for regular regional monitoring. Philip et al. [35] reported a high rate of β-lactam resistance in ICU patients with P. aeruginosa infection as compared with those of general hospitalised patients. Also reports by Juayang et al. [28] indicated that P. aeruginosa isolates in their study were most resistant to the fluoroquinolone levofloxacin while showing lower resistance to the aminoglycosides. These variations could be explained as differences in the regions of bacteria isolation. It is also worth noting that while majority of the isolates in Juayang et al. [28] study fell within an MAR index of less than 0.2, majority (91%) of those in the present investigation fell in MAR index that was greater than 0.2 with a mean of 0.43. This therefore means that with the bacteria isolates in this study having MAR index ≥ 0.2, as stipulated by Davies et al. [29], they were originating from a high risk source of contamination where several antibiotics are used. Earlier reports [37] had attributed the overuse or misuse of antibiotics to be one of the reasons of growing resistance among bacteria isolates. The region of the present study had been reported [38] to be a region of high use of non-prescription antibiotics and this could explain the reason for the obtained MAR index result among the isolates in this study. However, the view expressed by Yusuf et al. [16] is that resistance to antibiotics as seen in ICU cases in their study was linked to the duration of patient treatment with antibiotics. They indicated that a less than 4 d of treatment was not associated with the emergency of resistance while 15 d of treatment with antibiotic did result in anti-Pseudomonal drug resistance but with the exception of amikacin. In this study, an average high sensitivity rate was seen among the isolates to amikacin (82%), gentamicin (81%) and piperacillin/tazobactam (76%). These findings have been reported [28] and this phenomenon was explained to be due to clonal spread of P. aeruginosa. While noting amikacin to be the most effective drug in their study, Juayang et al. [28] did draw attention to the adverse effects of this antibiotic as had earlier been pointed out [32]. Thus piperacillin/tazobactam and gentamicin remain the drugs of choice also a view of other researchers [16,39,40]. As regards the carbapenems, this study shows a five year mean percentage resistance for both imipenem and meropenem to be 32.3% and 34.9% respectively. This is a disturbing development considering the fact that the carbapenems are last line antibiotic used in the treatment of Gram negative bacteria. Resistance to the carbapenems by P. aeruginosa has been reported by researchers [16,28] in other regions of the world. This is an alarming development needing urgent attention as had been pointed out [28]. However, Yusuf et al. [16] were of the view that P. aeruginosa resistance to the carbapenems would only evolve if patient treatment is for 4 days or more, a view that had been expressed by Lodise et al. [41]. This might explain the reason for the carbapenem resistance shown by the isolates in the present investigation. This generally highlights the need for the careful monitoring of extended use of antibiotics even in intensive care units. The question as regards treatment based on the source of the specimen is answered by the results in the Table 2. There is a noticeable variability between the source of specimen and susceptibility to antibiotics. Although Juayang et al. [28] were of the view that antibiotics are effective irrespective of the anatomical structure of isolation, this is not the case seen in the present investigation as there seemed to be a clear exhibition of resistance to antimicrobials based on the source of specimens. There will be for further and more detailed investigation on this aspect as no conclusions can be drawn on this.
In terms of age, majority (81.25%) of the patients were elderly pointing to a decrease in immunity as a result of age and that most (46.5%) of the isolates had been from sputum could mean they were from patients with pneumonia. Similar findings had been reported in a recent study by Juayang et al. [28]. They observed from a 5 year study that P. aeruginosa infection was prevalent among older patients and explained this to be as a result of age-associated reduced immunity. This is a view that had earlier been expressed by other researchers [42-44].
Conclusion
Generally, the present study has shown that P. aeruginosa can still be treated with anti-Pseudomonal drugs. There is however a growing resistance trend that needs to be watched. Also the resistance to antimicrobials as seen by the isolates from this region of study could be attributed to the fact that they are from a region of high antibiotic use. There is need for a continuous monitoring.
Acknowledgement
The researchers would like to thank Mr. Hani Hussain Al- Farhan for his technical assistance.
References
- Streeter K, Katouli M. Pseudomonas aeruginosa: a review of their pathogenesis and prevalence in clinical settings and the environment. Infect Epidemiol Med 2016; 2: 25-32.
- Coggan KA, Wolfgang MC. Global regulatory pathways and cross-talk control Pseudomonas aeruginosa environmental lifestyle and virulence phenotype. Curr Issues Mol Biol 2012; 14: 47-70.
- Pirnay JP, Matthijs S, Colak H, Chablain P, Bilocq F, Van Eldere J. Global Pseudomonas aeruginosa biodiversity as reflected in a Belgian river. Environ Microbiol 2005; 7: 969-980.
- Riou M, Carbonnelle S, Avrain L, Mesaros N, Pirnay JP, BilocqF. In vivo development of antimicrobial resistance in Pseudomonas aeruginosa strains isolated from the lower respiratory tract of intensive care unit patients with nosocomial pneumonia and receiving antipseudomonal therapy. Int J Antimicrob Agents 2010; 36: 513-522.
- Brown SP, Cornfort DM, Mideo N. Evolution of virulence in opportunistic pathogens: generalism, plasticity and control. Trends Microbiol 2012; 20: 336-342.
- Alshaiki JMM, Toweir AA. Prevalance Pseudomonas aeruginosa Among Libyan patients and its association with hospitals environment in Benghazi. J Med Microb Diagn 2017; 6: 257.
- Mittal R, Aggarwal S, Sharma S, Chhibber S, Harjai K. Urinary tract infections caused by Pseudomonas aeruginosa: a mini review. J Infect Public Health 2009; 2: 101-111.
- Fatima A, Naqvi SB, Khaliq SA, Perveen S, Jabeen S. Antimicrobial susceptibility pattern of clinical isolates of Pseudomonas aeruginosa isolated from patients of lower respiratory tract infections. Springer Plus 2012; 1: 70.
- Mansoor T, Musani MA, Khalid G, Kamal M. Pseudomonas aeruginosa in chronic suppurative otitis media: sensitivity spectrum against various antibiotics in Karachi. J Ayub Med Coll Abbottabad 2009; 21: 120-123.
- Kim M, Christley S, Khodarev NN, Fleming I, Huang Y, Chang E. Pseudomonas aeruginosa wound infection involves activation of its iron acquisition system in response to fascial contact. J Trauma Acute Care Surg 2015; 78: 823-829.
- Raffaele S, Raffaele G, Lucia B, Alessio R, Ugo F. Chronic wound infections: the role of Pseudomonas aeruginosaand Staphylococcus aureus. Expert Rev Anti Infect Ther 2015; 13: 605-613.
- Bhagirath AY, Li Y, Somayajula D, Dadashi M, Badr S, Duan K. Cystic fibrosis lung environment and Pseudomonas aeruginosa infection. BMC Pulm Med 2016; 16: 174.
- Fujitani S, Sun HY, Yu VL, Weingarten JA. Pneumonia due to Pseudomonas aeruginosa: part 1: epidemiology, clinical diagnosis and source. Chest 2011; 139: 909-919.
- ECDC. European Centre for Disease Prevention and Control. Point prevalence Survey of health care associated infections antimicrobial use in European acute care hospitals. Stockholm: ECDC 2013.
- Vikas CY, Vepada RK, Mahendra KJ, Khileshwar S. A study of antibiotic sensitivity pattern of Pseudomonas aeruginosa isolated from a tertiary care hospital in South Chhattisgarh. Int J Med Sci Public Health 2016; 6; 600-605.
- Yusuf E, Bruno VH, Walter V, Margareta I, Emiel G. Emergence of antimicrobial resistance to Pseudomonas aeruginosa in the intensive care unit: association with the duration of antibiotic exposure and mode of administration. Ann Intensive Care 2017; 7: 72.
- Yadav VC, Kiran VR, Jaiswal MK, Singh K. A study of antibiotic sensitivity pattern of Pseudomonas aeruginosa isolated from a tertiary care hospital in South Chhattisgarh. Int J Med Sci Public Health 2017; 6: 600-605.
- Rakesh MR, Govind LN, Kalpesh M, Rosy P, Kavu P, Vegad MM. Antibiotic resistance pattern in Pseudomonas aeruginosa species isolated at a tertiary care hospital, Ahmadabad. Natl J Med Res 2012; 2: 156-159.
- Mayhall CG. Nosocomial burn wound infection. In: Mayhall GC (ed). Hospital epidemiology and infection control. William and Wilkins Co, Baltimore, MD, USA 1996; 225-236.
- Javiya VA, Ghatak SB, Patel KR, Patel JA. Antibiotic susceptibility patterns of Pseudomonas aeruginosa at a tertiary care hospital in Gujarat, India. Indian J Pharmacol 2008; 40: 230-234.
- Cobos-Trigueros N, Sole M, Castro P, Torres JL. Acquisition of Pseudomonas aeruginosa and its resistance phenotypes in critically ill medical patients: role of colonization pressure and antibiotic exposure. Crit Care 2015; 19: 218.
- Samporn S, Chuntima T, Thitiya Y, Chertask D. Prevalence and antimicrobial susceptibility of pseudomonas aeruginosa mucoid and non-mucoid type. SE Asian J Trop Med 2004; 35: 893-894.
- Marilyn Porras-Gomez, Jose Vega-Baudrit, Santiago Nunez-Corrales. Overview of Multidrug-Resistant Pseudomonas aeruginosa and Novel Therapeutic Approaches. J Biomater Nanobiotechnol 2012; 3: 519-527.
- Ozer B, Tatman-Otkun M, Memis D, Otkun M. Characteristics of Pseudomonas aeruginosa Isolates from Intensive Care Unit. Cent Eur J Med 2009; 4: 156-163.
- Ishraq AAS, Israa AI, Hawrra WA. Pseudomonas aeruginosa; antibiotics susceptibility among patients of nosocomial infection in Hillah city, Iraq. Int J Cur Eng Tech 2015; 5: 1577-1579.
- Dotsch A, Becker T, Pommerenke C, Magnowska Z, Jansch L, Haussler S. Genomewide identification of genetic determinants of antimicrobial drug resistance in Pseudomonas aeruginosa. Antimicrob Agents Chemother 2009; 53: 2522-2531.
- Eva T, Rosa V, Germa’n B. Comparison of different methods of determining ß-lactam susceptibility in clinical strains of Pseudomonas aeruginosa. J Med Microbiol 2009; 58: 625-629.
- Juayang AC, Lim JPT, Bonifacio AFV, Lambot AVL, Millan SM. Five-year antimicrobial susceptibility of Pseudomonas aeruginosa from a local tertiary Hospital in Bacolod City, Philippines. Trop Med Infect Dis 2017; 2: 28.
- Collins CH, Lyne PM, Grange JM, Falkinham JO. III. Collins and Lynes Microbiological Methods (8th Edn.). Arnolds London UK 2004.
- Forbes B, Sahm D, Weissfeld A. Bailey and Scotts Diagnostic Microbiology (12th Edn.). Mosby Elsevier: St. Louis, MO, USA 2007.
- Clinical and Laboratory Standards Institute. Performance standards for antimicrobial susceptibility testing: Twenty Fifth Informational Supplement M100-S23; CLSI: Wayne, PA, USA 2015.
- Badger-Emeka L, Emeka PM, Dibua UME. Plasmid profile of the Nigerian strain of multi-drug resistant clinical isolates of Staphylococcus aureus. Afr J Biotechnol 2014; 13: 4148-4154.
- Christopher H, Alnassafi K, Honghuah H, Mark E, Peter B. Modulation of gene expression by Pseudomonas aeruginosa during chronic infection in the adult cystic fibrosis lung. Microbiology 2013; 159: 2354-2363.
- Subramani S, Vignesh A. MAR index study and MDR Character analysis of a few golden Staphylococci isolates. Asian J Pharm 2012; 2: 151-154.
- Philip DL, Daniel JW, Nancy DH. Antibacterial-resistant pseudomonas aeruginosa: clinical impact and complex regulation of chromosomally encoded resistance mechanisms. Clin Microbiol Rev 2009; 22: 582-610.
- Sedighi M, Safiri S, Pirouzi S, Jayasinghe H, Sepidarkish M, Fouladseresht H. Detection and determination of the antibiotic resistance patterns in Pseudomonas aeruginosa strains isolated from clinical specimens in hospitals of Isfahan, Iran, 2012. Scimetr 2015; 3: 21133.
- Berglund B. Environmental dissemination of antibiotic resistance genes and correlation to anthropogenic contamination with antibiotics. J Infect Ecol Epidemiol 2015; 5: 28564.
- Emeka PM, Al-Omar M, Khan TM. Public attitude and justification to purchase antibiotics in the Eastern region Al Ahsa of Saudi Arabia. Saudi Pharm J 2014; 2: 550-554.
- Antimicrobial Resistance Surveillance Reference Laboratory. Antimicrobial resistance surveillance program 2015 data summary report; Research Institute for Tropical Medicine, Department of Health: Manila, Philippines 2015.
- Pathmanathan SG, Samat NA, Mohamed R. Antimicrobial susceptibility of clinical isolates of Pseudomonas aeruginosa from a Malaysian hospital. MJMS 2009; 16: 27-32.
- Lodise TP, Miller C, Patel N, Graves J, McNutt LA. Identification of patients with Pseudomonas aeruginosa respiratory tract infections at greatest risk of infection with carbapenem-resistant isolates. Infect Control Hosp Epidemiol 2007; 28: 959-965.
- Juayang A, Maestral D, de los Reyes G, Acosido MA, Gallega C. Review on the antimicrobial resistance of pathogens from tracheal and endotracheal aspirates of patients with clinical manifestations of pneumonia in Bacolod City in 2013. Int J Bacteriol 2015; 942509.
- Yayan J, Ghebremedhin B, Rasche K. Antibiotic resistance of Pseudomonas aeruginosa in pneumonia at a single university hospital center in Germany over a 10-year period. PLoS One 2015; 10: 0139836.
- Scheld WM, Grayson ML, Hughes JM. Infections in long-term care facilities in emerging infections (9th Edn.). ASM Press: New Washington DC USA 2010; 287-288.